 -
Volume 60,
Issue 1,
2010
-
Volume 60,
Issue 1,
2010
Volume 60, Issue 1, 2010
- New Taxa
-
- Firmicutes And Related Organisms
-
-
Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov.
More LessAn obligately anaerobic, xylanolytic, extremely thermophilic bacterium, strain JW/HY-331T, was isolated from sheep faeces collected from a farm at the University of Georgia, USA. Cells of strain JW/HY-331T stained Gram-positive and were catalase-negative, non-motile rods. Single terminal endospores (0.4–0.6 μm in diameter) swelled the mother cell. Growth ranges were 44–77 °C (optimum 70 °C at pH70 °C 7.2) and pH70 °C 5.9–8.6 (optimum 7.2 at 70 °C). Salt tolerance was 0–2.0 % (w/v) NaCl. No growth was observed at or below 42 °C or at or above 79 °C or at pH70 °C 5.7 and below or 8.9 and above. In the presence of 0.3 % yeast extract and 0.1 % tryptone, strain JW/HY-331T utilized xylose, glucose, galactose, cellobiose, raffinose and xylan as carbon and energy sources, but not dextran, soluble potato starch, CM-cellulose, cellulose powder, casein or Casamino acids. Fermentation products from glucose were lactate, acetate, ethanol, CO2 and H2. The G+C content of the genomic DNA was 45.4 mol% (HPLC). Major cellular fatty acids were iso-C17 : 0, iso-C15 : 0 and anteiso-C17 : 0. No respiratory quinones were detected. The cell-wall structure was a single layer (Gram-type positive) of the peptidoglycan type A1γ; the cell-wall sugars were galactose and mannose. Based on 16S rRNA gene sequence analysis, ‘Catabacter hongkongensis’ HKU16 (85.4 % similarity), Caloramator fervidus ATCC 43204T (84.2 %) and Caloranaerobacter azorensis MV1087T (83.4 %) were the closest relatives, but they were only distantly related to strain JW/HY-331T. On the basis of physiological, chemotaxonomic and phylogenetic data, isolate JW/HY-331T (=DSM 21659T =ATCC BAA-1711T) is proposed as the type strain of Caldicoprobacter oshimai gen. nov., sp. nov., placed in Caldicoprobacteraceae fam. nov. within the order Clostridiales of the phylum Firmicutes.
-
-
-
Streptococcus porci sp. nov., isolated from swine sources
More LessTwo unidentified Gram-positive, catalase-negative, coccus-shaped organisms were recovered from pigs and subjected to a polyphasic taxonomic analysis. Based on cellular morphology and biochemical criteria, the isolates were tentatively assigned to the genus Streptococcus, although the organisms did not appear to correspond to any recognized species. Comparative 16S rRNA gene sequence studies confirmed this identification and showed that the nearest phylogenetic relatives of the unknown cocci were Streptococcus plurextorum 1956-02T and Streptococcus suis NCTC 10234T (97.9 and 96.0 % 16S rRNA gene sequence similarity, respectively). The new isolates were related most closely to S. suis CIP 103217T based on rpoB gene sequence analysis (<8 % sequence divergence). DNA–DNA pairing studies showed that one of the unidentified strains (2923-03T) displayed DNA relatedness values of 26.6 and 27.2 % with S. plurextorum CECT 7308T and S. suis NCTC 10234T, respectively. On the basis of phenotypic and phylogenetic evidence, it is proposed that the unknown isolates from pigs be classified in the genus Streptococcus as members of Streptococcus porci sp. nov., with the type strain 2923-03T (=CECT 7374T =CCUG 55896T).
-
-
-
Lactobacillus equicursoris sp. nov., isolated from the faeces of a thoroughbred racehorse
More LessWe previously isolated five strains of putative lactobacilli from the faeces of a thoroughbred horse (a 4-year-old male). Of the five strains, four were identified as members of existing Lactobacillus species; however, sequence analysis of the 16S rRNA gene revealed that the fifth isolate, DI70T, showed approximately 97 % identity (1325/1366 bp) with the type strain of Lactobacillus delbrueckii. Therefore, we considered the possibility that DI70T represents a novel species of the genus Lactobacillus. Cells of strain DI70T were Gram-stain-positive, catalase-negative, non-spore-forming, non-motile rods. In phylogenetic trees constructed on the basis of 16S rRNA gene sequences, strain DI70T formed a subcluster in the L. delbrueckii phylogenetic group and was closely related to L. delbrueckii, Lactobacillus crispatus and Lactobacillus jensenii. However, analysis of DNA–DNA relatedness showed that DI70T was genetically distinct from its phylogenetic relatives. The isolate also exhibited distinct biochemical and physiological characteristics when compared with its phylogenetic relatives. It required anaerobic conditions for growth on agar medium. The results indicate that isolate DI70T indeed represents a novel species of the genus Lactobacillus, for which we propose the name Lactobacillus equicursoris sp. nov. The type strain is DI70T (=JCM 14600T =DSM 19284T).
-
-
-
Paenibacillus riograndensis sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Triticum aestivum
More LessA bacterial strain designated SBR5T was isolated from the rhizosphere of Triticum aestivum. A phylogenetic analysis based on the 16S rRNA gene sequence placed the isolate within the genus Paenibacillus, being most closely related to Paenibacillus graminis RSA19T (98.1 % similarity). The isolate was a Gram-reaction-variable, motile, facultatively anaerobic bacterium, with spores in a terminal position in cells. Starch was utilized and dihydroxyacetone and catalase were produced. Strain SBR5T displayed plant-growth-promoting rhizobacteria characteristics: the ability to fix nitrogen and to produce siderophores and indole-3-acetic acid. The DNA G+C content was 55.1 mol%. Chemotaxonomic analysis of the isolated strain revealed that MK-7 was the predominant menaquinone, while the major fatty acid was anteiso-C15 : 0. DNA–DNA hybridization values between strain SBR5T and P. graminis RSA19T, Paenibacillus odorifer TOD45T and Paenibacillus borealis KK19T were 43, 35 and 28 %, respectively. These DNA relatedness data and the results of phylogenetic and phenotypic analyses showed that strain SBR5T should be considered as the nitrogen-fixing type strain of a novel species of the genus Paenibacillus, for which the name Paenibacillus riograndensis sp. nov. is proposed. The type strain is SBR5T (=CCGB 1313T =CECT 7330T).
-
-
-
Lactobacillus similis sp. nov., isolated from fermented cane molasses
More LessThe taxonomic position of strain JCM 2765T isolated from fermented cane molasses in Thailand was reinvestigated. Strain JCM 2765T was originally identified as representing Lactobacillus buchneri on the basis of biochemical and physiological characteristics. In the present study, 16S rRNA gene sequence analysis of strain JCM 2765T demonstrated a low level of similarity with the type strain of L. buchneri (92.5 %) and high levels with those of Lactobacillus collinoides (97.6 %) and Lactobacillus paracollinoides (98.0 %). Ribotyping was applied to investigate the relationships between strain JCM 2765T, L. collinoides and L. paracollinoides. The dendrogram based on ribotyping patterns showed one cluster for six strains of L. paracollinoides, and that strain JCM 2765T and L. collinoides JCM 1123T were each independent. Based on additional phenotypic findings and DNA–DNA hybridization results, strain JCM 2765T is considered to represent a novel species of the genus Lactobacillus, for which the name Lactobacillus similis sp. nov. is proposed. The type strain is JCM 2765T (=LMG 23904T).
-
- Other Bacteria
-
-
Thermodesulfatator atlanticus sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent
More LessA novel, strictly anaerobic, thermophilic, sulfate-reducing bacterium, designated strain AT1325T, was isolated from a deep-sea hydrothermal vent at the Rainbow site on the Mid-Atlantic Ridge. This strain was subjected to a polyphasic taxonomic analysis. Cells were Gram-negative motile rods (approximately 2.4×0.6 μm) with a single polar flagellum. Strain AT1325T grew at 55–75 °C (optimum, 65–70 °C), at pH 5.5–8.0 (optimum, 6.5–7.5) and in the presence of 1.5–4.5 % (w/v) NaCl (optimum, 2.5 %). Cells grew chemolithoautotrophically with H2 as an energy source and
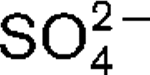 as an electron acceptor. Alternatively, the novel isolate was able to use methylamine, peptone or yeast extract as carbon sources. The dominant fatty acids (>5 % of the total) were C16 : 0, C18 : 1
ω7c, C18 : 0 and C19 : 0 cyclo ω8c. The G+C content of the genomic DNA of strain AT1325T was 45.6 mol%. Phylogenetic analyses based on 16S rRNA gene sequences placed strain AT1325T within the family Thermodesulfobacteriaceae, in the bacterial domain. Comparative 16S rRNA gene sequence analysis indicated that strain AT1325T belonged to the genus Thermodesulfatator, sharing 97.8 % similarity with the type strain of Thermodesulfatator indicus, the unique representative species of this genus. On the basis of the data presented, it is suggested that strain AT1325T represents a novel species of the genus Thermodesulfatator, for which the name Thermodesulfatator atlanticus sp. nov. is proposed. The type strain is AT1325T (=DSM 21156T=JCM 15391T).
as an electron acceptor. Alternatively, the novel isolate was able to use methylamine, peptone or yeast extract as carbon sources. The dominant fatty acids (>5 % of the total) were C16 : 0, C18 : 1
ω7c, C18 : 0 and C19 : 0 cyclo ω8c. The G+C content of the genomic DNA of strain AT1325T was 45.6 mol%. Phylogenetic analyses based on 16S rRNA gene sequences placed strain AT1325T within the family Thermodesulfobacteriaceae, in the bacterial domain. Comparative 16S rRNA gene sequence analysis indicated that strain AT1325T belonged to the genus Thermodesulfatator, sharing 97.8 % similarity with the type strain of Thermodesulfatator indicus, the unique representative species of this genus. On the basis of the data presented, it is suggested that strain AT1325T represents a novel species of the genus Thermodesulfatator, for which the name Thermodesulfatator atlanticus sp. nov. is proposed. The type strain is AT1325T (=DSM 21156T=JCM 15391T).
-
- Proteobacteria
-
-
Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.)
More LessA facultatively chemolithoautotrophic, thiosulfate-oxidizing, Gram-negative, aerobic, motile, rod-shaped bacterial strain, designated ATSB16T, was isolated from rhizosphere soils of sesame (Sesamum indicum L.). 16S rRNA gene sequence analysis demonstrated that this strain was closely related to Pandoraea pnomenusa LMG 18087T (96.7 % similarity), P. pulmonicola LMG 18016T (96.5 %), P. apista LMG 16407T (96.2 %), P. norimbergensis LMG 18379T (96.1 %) and P. sputorum LMG 18819T (96.0 %). Strain ATSB16T shared 96.0–96.4 % sequence similarity with four unnamed genomospecies of Pandoraea. The major cellular fatty acids of the strain ATSB16T were C17 : 0 cyclo (33.0 %) and C16 : 0 (30.6 %). Q-8 was the predominant respiratory quinone. The major polar lipids were phosphatidylmethylethanolamine, diphosphatidylglycerol, phosphatidylethanolamine and two unidentified aminophospholipids. Hydroxyputrescine and putrescine were the predominant polyamines. The genomic DNA G+C content of the strain was 64.0 mol%. On the basis of the results obtained from this study, strain ATSB16T represents a novel species of the genus Pandoraea, for which the name Pandoraea thiooxydans sp. nov. is proposed. The type strain is ATSB16T (=KACC 12757T =LMG 24779T).
-
-
-
Basfia succiniciproducens gen. nov., sp. nov., a new member of the family Pasteurellaceae isolated from bovine rumen
More LessGram-negative, coccoid, non-motile bacteria that are catalase-, urease- and indole-negative, facultatively anaerobic and oxidase-positive were isolated from the bovine rumen using an improved selective medium for members of the Pasteurellaceae. All strains produced significant amounts of succinic acid under anaerobic conditions with glucose as substrate. Phenotypic characterization and multilocus sequence analysis (MLSA) using 16S rRNA, rpoB, infB and recN genes were performed on seven independent isolates. All four genes showed high sequence similarity to their counterparts in the genome sequence of the patent strain MBEL55E, but less than 95 % 16S rRNA gene sequence similarity to any other species of the Pasteurellaceae. Genetically these strains form a very homogeneous group in individual as well as combined phylogenetic trees, clearly separated from other genera of the family from which they can also be separated based on phenotypic markers. Genome relatedness as deduced from the recN gene showed high interspecies similarities, but again low similarity to any of the established genera of the family. No toxicity towards bovine, human or fish cells was observed and no RTX toxin genes were detected in members of the new taxon. Based on phylogenetic clustering in the MLSA analysis, the low genetic similarity to other genera and the phenotypic distinction, we suggest to classify these bovine rumen isolates as Basfia succiniciproducens gen. nov., sp. nov. The type strain is JF4016T (=DSM 22022T =CCUG 57335T).
-
-
-
Salinarimonas rosea gen. nov., sp. nov., a new member of the α-2 subgroup of the Proteobacteria
More LessA Gram-negative, rod-shaped, facultatively anaerobic, halotolerant bacterial strain, designated YIM YD3T, was isolated from a salt mine in Yunnan, south-west China. The taxonomy of strain YIM YD3T was investigated by a polyphasic approach. Strain YIM YD3T was motile, formed pink colonies and was positive for catalase and oxidase activities. Q-10 was the predominant respiratory ubiquinone. The major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylmethylethanolamine, phosphatidylcholine and two unknown phospholipids. The major fatty acids (>10 % of total fatty acids) were C18 : 1 ω7c, C18 : 1 ω9c, C16 : 0 and C19 : 0 cyclo ω8c. The DNA G+C content was 71.8 mol%. Phylogenetic analysis based on 16S rRNA gene sequence comparisons showed that the isolate formed a distinct line within a clade containing the genera Balneimonas, Bosea, Chelatococcus and Microvirga in the order Rhizobiales, with highest levels of 16S RNA gene sequence similarity to the type strain of Balneimonas flocculans (93.5 %). On the basis of phenotypic, chemotaxonomic and phylogenetic data, strain YIM YD3T represents a novel species in a new genus, for which the name Salinarimonas rosea gen. nov., sp. nov. is proposed, with strain YIM YD3T (=KCTC 22346T=CCTCC AA208038T) as the type strain.
-
-
-
Aeromonas fluvialis sp. nov., isolated from a Spanish river
More LessA Gram-stain-negative, facultatively anaerobic bacterial strain, designated 717T, was isolated from a water sample collected from the Muga river, Girona, north-east Spain. Preliminary analysis of the 16S rRNA gene sequence showed that this strain belonged to the genus Aeromonas, the nearest species being Aeromonas veronii (99.5 % similarity, with seven different nucleotides). A polyphasic study based on a multilocus phylogenetic analysis of five housekeeping genes (gyrB, rpoD, recA, dnaJ and gyrA; 3684 bp) showed isolate 717T to be an independent phylogenetic line, with Aeromonas sobria, Aeromonas veronii and Aeromonas allosaccharophila as the closest neighbour species. DNA–DNA reassociation experiments and phenotypic analysis identified that strain 717T represents a novel species, for which the name Aeromonas fluvialis sp. nov. is proposed, with type strain 717T (=CECT 7401T =LMG 24681T).
-
-
-
Psychromonas boydii sp. nov., a gas-vacuolate, psychrophilic bacterium isolated from an Arctic sea-ice core
More LessA gas-vacuolate bacterium, strain 174T, was isolated from a sea-ice core collected from Point Barrow, Alaska, USA. Comparative analysis of 16S rRNA gene sequences showed that this bacterium was most closely related to Psychromonas ingrahamii 37T, with a similarity of >99 %. However, strain 174T could be clearly distinguished from closely related species by DNA–DNA hybridization; relatedness values determined by two different methods between strain 174T and P. ingrahamii 37T were 58.4 and 55.7 % and those between strain 174T and Psychromonas antarctica DSM 10704T were 46.1 and 33.1 %, which are well below the 70 % level used to define a distinct species. Phenotypic analysis, including cell size (strain 174T is the largest member of the genus Psychromonas, with rod-shaped cells, 8–18 μm long), further differentiated strain 174T from other members of the genus Psychromonas. Strain 174T could be distinguished from its closest relative, P. ingrahamii, by its utilization of d-mannose and d-xylose as sole carbon sources, its ability to ferment myo-inositol and its inability to use fumarate and glycerol as sole carbon sources. In addition, strain 174T contained gas vacuoles of two distinct morphologies and grew at temperatures ranging from below 0 to 10 °C and its optimal NaCl concentration for growth was 3.5 %. The DNA G+C content was 40 mol%. Whole-cell fatty acid analysis showed that 16 : 1ω7c and 16 : 0 comprised 44.9 and 26.4 % of the total fatty acid content, respectively. The name Psychromonas boydii sp. nov. is proposed for this novel species, with strain 174T (=DSM 17665T =CCM 7498T) as the type strain.
-
-
-
Taxonomic study of Marinomonas strains isolated from the seagrass Posidonia oceanica, with descriptions of Marinomonas balearica sp. nov. and Marinomonas pollencensis sp. nov.
More LessNovel aerobic, Gram-negative bacteria with DNA G+C contents below 50 mol% were isolated from the culturable microbiota associated with the Mediterranean seagrass Posidonia oceanica. 16S rRNA gene sequence analyses revealed that they belong to the genus Marinomonas. Strain IVIA-Po-186 is a strain of the species Marinomonas mediterranea, showing 99.77 % 16S rRNA gene sequence similarity with the type strain, MMB-1T, and sharing all phenotypic characteristics studied. This is the first description of this species forming part of the microbiota of a marine plant. A second strain, designated IVIA-Po-101T, was closely related to M. mediterranea based on phylogenetic studies. However, it differed in characteristics such as melanin synthesis and tyrosinase, laccase and antimicrobial activities. In addition, strain IVIA-Po-101T was auxotrophic and unable to use acetate. IVIA-Po-101T shared 97.86 % 16S rRNA gene sequence similarity with M. mediterranea MMB-1T, but the level of DNA–DNA relatedness between the two strains was only 10.3 %. On the basis of these data, strain IVIA-Po-101T is considered to represent a novel species of the genus Marinomonas, for which the name Marinomonas balearica sp. nov. is proposed. The type strain is IVIA-Po-101T (=CECT 7378T =NCIMB 14432T). A third novel strain, IVIA-Po-185T, was phylogenetically distant from all recognized Marinomonas species. It shared the highest 16S rRNA gene sequence similarity (97.4 %) with the type strain of Marinomonas pontica, but the level of DNA–DNA relatedness between the two strains was only 14.5 %. A differential chemotaxonomic marker of this strain in the genus Marinomonas is the presence of the fatty acid C17 : 0 cyclo. Strain IVIA-Po-185T is thus considered to represent a second novel species of the genus, for which the name Marinomonas pollencensis sp. nov. is proposed. The type strain is IVIA-Po-185T (=CECT 7375T =NCIMB 14435T). An emended description of the genus Marinomonas is given based on the description of these two novel species, as well as other Marinomonas species described after the original description of the genus.
-
-
-
Shinella fusca sp. nov., isolated from domestic waste compost
More LessA bacterium, designated strain DC-196T, isolated from kitchen refuse compost was analysed by using a polyphasic approach. Strain DC-196T was characterized as a Gram-negative short rod that was catalase- and oxidase-positive, and able to grow at 10–40 °C, pH 6–9 and in NaCl concentrations as high as 3 %. Chemotaxonomically, C18 : 1 was observed to be the predominant cellular fatty acid and ubiquinone 10 (Q10) was the predominant respiratory quinone. The G+C content of the genomic DNA was determined to be 66 mol%. On the basis of the genotypic, phenotypic and chemotaxonomic characteristics, strain DC-196T was assigned to the genus Shinella, although with distinctive features. At the time of writing, 16S rRNA gene sequence similarities of 97.6–96.8 % and the low DNA–DNA hybridization values of 38.2–32.2 % with the type strains of the three recognized Shinella species confirmed that strain DC-196T represents a novel species of the genus, for which the name Shinella fusca sp. nov. is proposed (type strain DC-196T=CCUG 55808T=LMG 24714T).
-
-
-
Polynucleobacter cosmopolitanus sp. nov., free-living planktonic bacteria inhabiting freshwater lakes and rivers
More LessFive heterotrophic, aerobic, catalase- and oxidase-positive, non-motile strains were characterized from freshwater habitats located in Austria, France, Uganda, P. R. China and New Zealand. The strains shared 16S rRNA gene similarities of ≥99.3 %. The novel strains grew on NSY medium over a temperature range of 10–35 °C (two strains also grew at 5 °C and one strain grew at 38 °C) and a NaCl tolerance range of 0.0–0.3 % (four strains grew up to 0.5 % NaCl). The predominant fatty acids were C16 : 0, C18 : 1 ω7c, C12 : 0 3-OH, and summed feature 3 (including C16 : 1 ω7c). The DNA G+C content of strain MWH-MoIso2T was 44.9 mol%. Phylogenetic analysis of 16S rRNA gene sequences demonstrated that the five new strains formed a monophyletic cluster closely related to Polynucleobacter necessarius (96–97 % sequence similarity). This cluster also harboured other isolates as well as environmental sequences which have been obtained from several habitats. Investigations with taxon-specific FISH probes demonstrated that the novel bacteria dwell as free-living, planktonic cells in freshwater systems. Based on the revealed phylogeny and pronounced chemotaxonomic differences to P. necessarius (presence of >7 % C12 : 0 3-OH and absence of C12 : 0 and C12 : 0 2-OH), the new strains are suggested to represent a novel species, for which the name Polynucleobacter cosmopolitanus sp. nov. is proposed. The type strain is MWH-MoIso2T (=DSM 21490T=CIP 109840T=LMG 25212T). The novel species belongs to the minority of described species of free-living bacteria for which both in situ data from their natural environments and culture-based knowledge are available.
-
-
-
Silvimonas iriomotensis sp. nov. and Silvimonas amylolytica sp. nov., new members of the class Betaproteobacteria isolated from the subtropical zone in Japan
More LessThe taxonomic positions of two bacterial strains, ir6-1T and ir6-4T, isolated from soil collected in Iriomote Island in Japan, were determined by using a polyphasic taxonomic approach. The strains were facultatively anaerobic, motile and Gram-stain-negative rods and their optimum pH for growth was pH 4.0. Their major respiratory quinone was ubiquinone-8 and the predominant cellular fatty acids were C14 : 0, C16 : 0, C16 : 1 ω7c and C18 : 1 ω7c. The G+C content of the genomic DNA of ir6-1T and ir6-4T was 59.9 and 57.5 mol%, respectively. Phylogenetic analysis based on 16S rRNA gene sequences showed that the strains clustered with the genus Silvimonas in the class Betaproteobacteria. DNA–DNA similarities were lower than 53 % among ir6-1T, ir6-4T and Silvimonas terrae NBRC 100961T, and these strains could be differentiated from each other by several phenotypic characters. Based on these results, we propose the emendation of the genus Silvimonas and inclusion of two novel species, Silvimonas iriomotensis sp. nov. (type strain ir6-1T=NBRC 103188T =CGMCC 1.8859T =KCTC 22513T) and Silvimonas amylolytica sp. nov. (ir6-4T =NBRC 103189T =CGMCC 1.8860T =KCTC 22514T).
-
-
-
Mangrovibacter plantisponsor gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from a mangrove-associated wild rice (Porteresia coarctata Tateoka)
More LessA facultatively anaerobic, nitrogen-fixing bacterium, strain MSSRF40T, was isolated from roots of mangrove-associated wild rice (Porteresia coarctata Tateoka). On the basis of 16S rRNA gene sequence similarities, strain MSSRF40T was shown to belong to the family Enterobacteriaceae, most closely related to Cronobacter muytjensii E603T (97.2 % sequence similarity), Enterobacter cloacae subsp. dissolvens LMG 2683T (97.1 %), E. radicincitans D5/23T (97.1 %) and E. ludwigii EN-119T (97.0 %). Sequence analysis of rpoB, gyrB and hsp60 genes showed that strain MSSRF40T had relatively low sequence similarity (<91, <84 and <90 %) to recognized species of different genera of the family Enterobacteriaceae and formed an independent phyletic lineage in all phylogenetic analyses using the 16S rRNA, rpoB, gyrB and hsp60 genes, clearly indicating that strain MSSRF40T could not be affiliated to any of the recognized genera within the family Enterobacteriaceae. The dominant cellular fatty acids were C16 : 0, C16 : 1 ω7c and/or iso-C15 : 0 2-OH and C18 : 1 ω7c, similar to those of other members of the Enterobacteriaceae. The DNA G+C content was 50.1 mol%. Phylogenetic distinctiveness and phenotypic differences from its phylogenetic neighbours indicated that strain MSSRF40T represents a novel species and genus within the family Enterobacteriaceae, for which the name Mangrovibacter plantisponsor gen. nov., sp. nov. is proposed. The type strain of Mangrovibacter plantisponsor is strain MSSRF40T (=LMG 24236T =DSM 19579T).
-
-
-
Jannaschia seohaensis sp. nov., isolated from a tidal flat sediment
More LessA Gram-negative, motile and pleomorphic bacterial strain, SMK-146T, was isolated from a tidal flat sediment of the Yellow Sea, Korea, and its taxonomic position was investigated. Strain SMK-146T grew optimally at pH 7.0–8.0 and 30 °C. It contained Q-10 as the predominant ubiquinone and C18 : 1 ω7c and 11-methyl C18 : 1 ω7c as the major fatty acids. The major polar lipids were phosphatidylcholine, phosphatidylglycerol and phosphatidylethanolamine. The DNA G+C content was 68.4 mol%. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain SMK-146T belongs to the genus Jannaschia. Strain SMK-146T exhibited 16S rRNA gene sequence similarity values of 95.3–97.0 % to the type strains of the five recognized Jannaschia species. The mean DNA–DNA relatedness value between strain SMK-146T and Jannaschia seosinensis KCCM 42114T, the closest phylogenetic neighbour, was 17 %. Differential phenotypic properties also revealed that strain SMK-146T differs from the recognized Jannaschia species. On the basis of phenotypic, phylogenetic and genetic data, strain SMK-146T represents a novel species of the genus Jannaschia, for which the name Jannaschia seohaensis sp. nov. is proposed. The type strain is SMK-146T (=KCTC 22172T =CCUG 55326T).
-
-
-
Gaetbulicola byunsanensis gen. nov., sp. nov., isolated from tidal flat sediment
More LessA Gram-negative, non-motile and pleomorphic bacterial strain, SMK-114T, which belongs to the class Alphaproteobacteria, was isolated from a tidal flat sample collected in Byunsan, Korea. Strain SMK-114T grew optimally at pH 7.0–8.0 and 25–30 °C and in the presence of 2 % (w/v) NaCl. A neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showed that strain SMK-114T formed a cluster with Octadecabacter species, with which it exhibited 16S rRNA gene sequence similarity values of 95.2–95.4 %. This cluster was part of the clade comprising Thalassobius species with a bootstrap resampling value of 76.3 %. Strain SMK-114T exhibited 16S rRNA gene sequence similarity values of 95.1–96.3 % to members of the genus Thalassobius. It contained Q-10 as the predominant ubiquinone and C18 : 1 ω7c as the major fatty acid. The DNA G+C content was 60.0 mol%. On the basis of phenotypic, chemotaxonomic and phylogenetic data, strain SMK-114T is considered to represent a novel species in a new genus for which the name Gaetbulicola byunsanensis gen. nov., sp. nov. is proposed. The type strain of Gaetbulicola byunsanensis is SMK-114T (=KCTC 22632T =CCUG 57612T).
-
-
-
Psychrobacter piscatorii sp. nov., a psychrotolerant bacterium exhibiting high catalase activity isolated from an oxidative environment
More LessA Gram-negative, non-motile, psychrotolerant bacterium exhibiting high catalase activity, designated strain T-3-2T, was isolated from a drain of a fish-processing plant. Its catalase activity was 12 000 U (mg protein)−1, much higher than the activity of the other Psychrobacter strains tested. The strain grew at 0–30 °C and in the presence of 0–12 % NaCl. The predominant isoprenoid quinone was ubiquinone-8 (Q-8), and C16 : 1 ω9c and C18 : 1 ω9c were the predominant cellular fatty acids. The DNA G+C content of strain T-3-2T was 43.9 mol%. 16S rRNA gene sequence phylogeny suggested that strain T-3-2T is a member of the genus Psychrobacter, with the closest relatives being the type strains of Psychrobacter nivimaris (99.2 % similarity), P. aquimaris (98.7 %) and P. proteolyticus (98.5 %). DNA–DNA hybridization showed less than 65 % relatedness with these strains. A phylogenetic tree based on gyrB gene sequences was more reliable, with higher bootstrap values than the 16S rRNA gene sequence-based tree. The result also differentiated the isolate from previously reported Psychrobacter species. Owing to the significant differences in phenotypic and chemotaxonomic characteristics and the phylogenetic and DNA–DNA relatedness data, the isolate merits classification within a novel species, for which the name Psychrobacter piscatorii sp. nov. is proposed. The type strain is T-3-2T (=JCM 15603T =NCIMB 14510T).
-
-
-
Thalassobaculum salexigens sp. nov., a new member of the family Rhodospirillaceae from the NW Mediterranean Sea, and emended description of the genus Thalassobaculum
More LessA novel Gram-negative bacteria, named CZ41_10aT, was isolated from coastal surface waters of the north-western Mediterranean Sea. Cells were motile, pleomorphic rods, 1.6 μm long and 0.7 μm wide and formed cream colonies on marine agar medium. The G+C content of the genomic DNA was 65 mol%. Phylogenetic analysis of 16S rRNA gene sequences placed the new isolate in the genus Thalassobaculum, a member of the family Rhodospirillaceae, class Alphaproteobacteria. Unlike Thalassobaculum litoreum CL-GR58T, its closest relative, strain CZ41_10aT was unable to grow anaerobically and did not exhibit nitrate reductase activity. On the basis of DNA–DNA hybridization, fatty acid content and physiological and biochemical characteristics, this isolate represents a novel species for which the name Thalassobaculum salexigens sp. nov. is proposed. The type strain is CZ41_10aT (=DSM 19539T=CIP 109064T=MOLA 84T). An emended description of the genus Thalassobaculum is also given.
-
Volumes and issues
-
Volume 74 (2024)
-
Volume 73 (2023)
-
Volume 72 (2022 - 2023)
-
Volume 71 (2020 - 2021)
-
Volume 70 (2020)
-
Volume 69 (2019)
-
Volume 68 (2018)
-
Volume 67 (2017)
-
Volume 66 (2016)
-
Volume 65 (2015)
-
Volume 64 (2014)
-
Volume 63 (2013)
-
Volume 62 (2012)
-
Volume 61 (2011)
-
Volume 60 (2010)
-
Volume 59 (2009)
-
Volume 58 (2008)
-
Volume 57 (2007)
-
Volume 56 (2006)
-
Volume 55 (2005)
-
Volume 54 (2004)
-
Volume 53 (2003)
-
Volume 52 (2002)
-
Volume 51 (2001)
-
Volume 50 (2000)
-
Volume 49 (1999)
-
Volume 48 (1998)
-
Volume 47 (1997)
-
Volume 46 (1996)
-
Volume 45 (1995)
-
Volume 44 (1994)
-
Volume 43 (1993)
-
Volume 42 (1992)
-
Volume 41 (1991)
-
Volume 40 (1990)
-
Volume 39 (1989)
-
Volume 38 (1988)
-
Volume 37 (1987)
-
Volume 36 (1986)
-
Volume 35 (1985)
-
Volume 34 (1984)
-
Volume 33 (1983)
-
Volume 32 (1982)
-
Volume 31 (1981)
-
Volume 30 (1980)
-
Volume 29 (1979)
-
Volume 28 (1978)
-
Volume 27 (1977)
-
Volume 26 (1976)
-
Volume 25 (1975)
-
Volume 24 (1974)
-
Volume 23 (1973)
-
Volume 22 (1972)
-
Volume 21 (1971)
-
Volume 20 (1970)
-
Volume 19 (1969)
-
Volume 18 (1968)
-
Volume 17 (1967)
-
Volume 16 (1966)
-
Volume 15 (1965)
-
Volume 14 (1964)
-
Volume 13 (1963)
-
Volume 12 (1962)
-
Volume 11 (1961)
-
Volume 10 (1960)
-
Volume 9 (1959)
-
Volume 8 (1958)
-
Volume 7 (1957)
-
Volume 6 (1956)
-
Volume 5 (1955)
-
Volume 4 (1954)
-
Volume 3 (1953)
-
Volume 2 (1952)
-
Volume 1 (1951)
Most Read This Month


