Collections
Browse our collections – bringing together peer-reviewed content from across the Society’s publishing platform on a range of hot topics and subject areas.
1 - 20 of 56 results
-
-
Anaerobe
More Less
Anaerobic clinical microbiology remains a challenge due to specialist culture requirements, coupled with the increase in and spread of antimicrobial resistance. The normal human microbiota is primarily composed of anaerobic bacteria, and is now recognised as a source of life-threatening anaerobic infection. More recent metataxonomic and metagenomic sequencing has extended interest in the potential role of the microbiota in a plethora of other aspects of human health, from obesity to mental health. In addition, the successful use of faecal microbiota transplants for the treatment of clostridial infection raises potential unchartered long-term consequences and possibilities.
In conjunction with Anaerobe 2019 and Anaerobe 2021, this collection will provide scientific insights into the future impact of anaerobic bacteria in human health and disease, addressing the implications of recent microbiota studies as well as the continued threat of emerging and re-emerging anaerobic infection.
This collection is open for submissions – please submit your article here, stating that your manuscript is part of the Anaerobe collection.
-
-
-
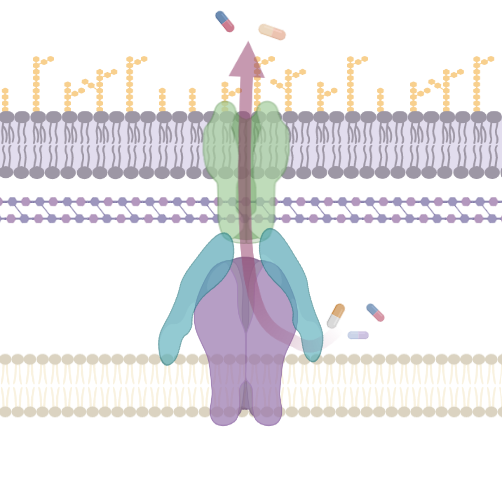
Antimicrobial Efflux
More Less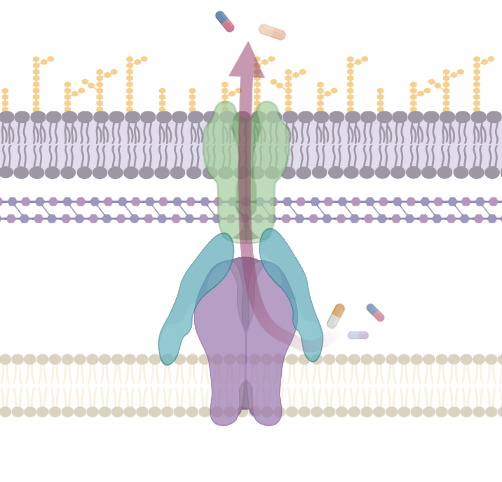
Bacterial efflux pumps are a complex and diverse set of membrane proteins responsible for transport of substrates between cellular compartments or to the outside of bacterial cells. They are important mediators of antimicrobial resistance and are critical for many bacterial pathogens to cause infection and form biofilm. This collection has been put together to mark two very special anniversaries. It is 40 years since George and Levy (1983) published the discovery of the first 'energy-dependent efflux system' conferring resistance to Tetracycline and 30 years since the first description of the pumps from the RND family (Poole et al., 1993 and Ma et al., 1993). In this time our understanding of efflux function, physiology and structure have been transformed. This special collection is guest edited by Drs Jessica Blair and Ayush Kumar and aims to highlight the diversity of efflux pump research and the most recent advances in this fascinating field. We also invite articles describing advances in methodology that aid the characterization of efflux pumps. This collection is open for submissions in Microbiology. Upon submission, please indicate that your manuscript is to be considered for the Antimicrobial Efflux collection.
-
-
-
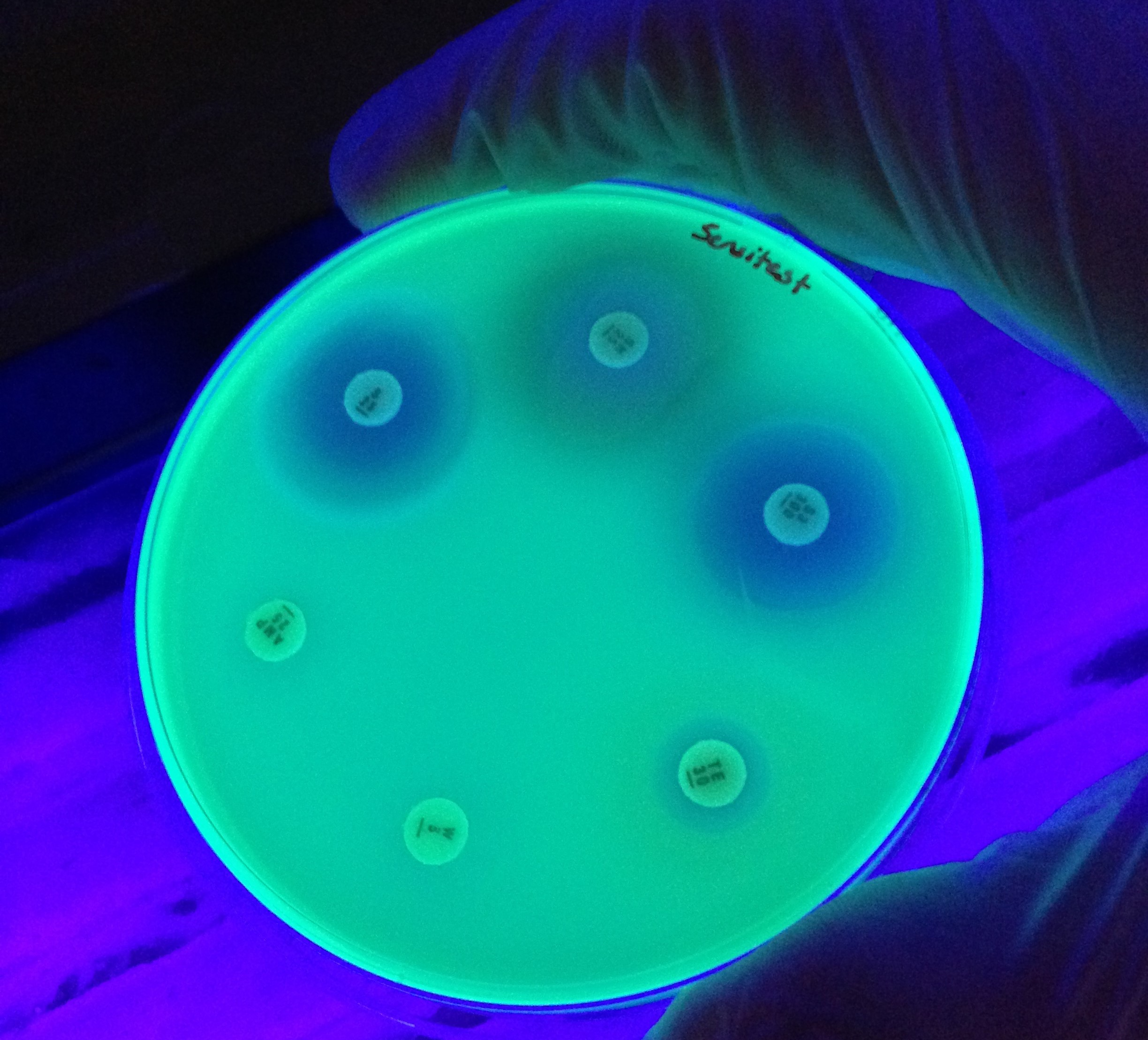
Antimicrobial Resistance
More Less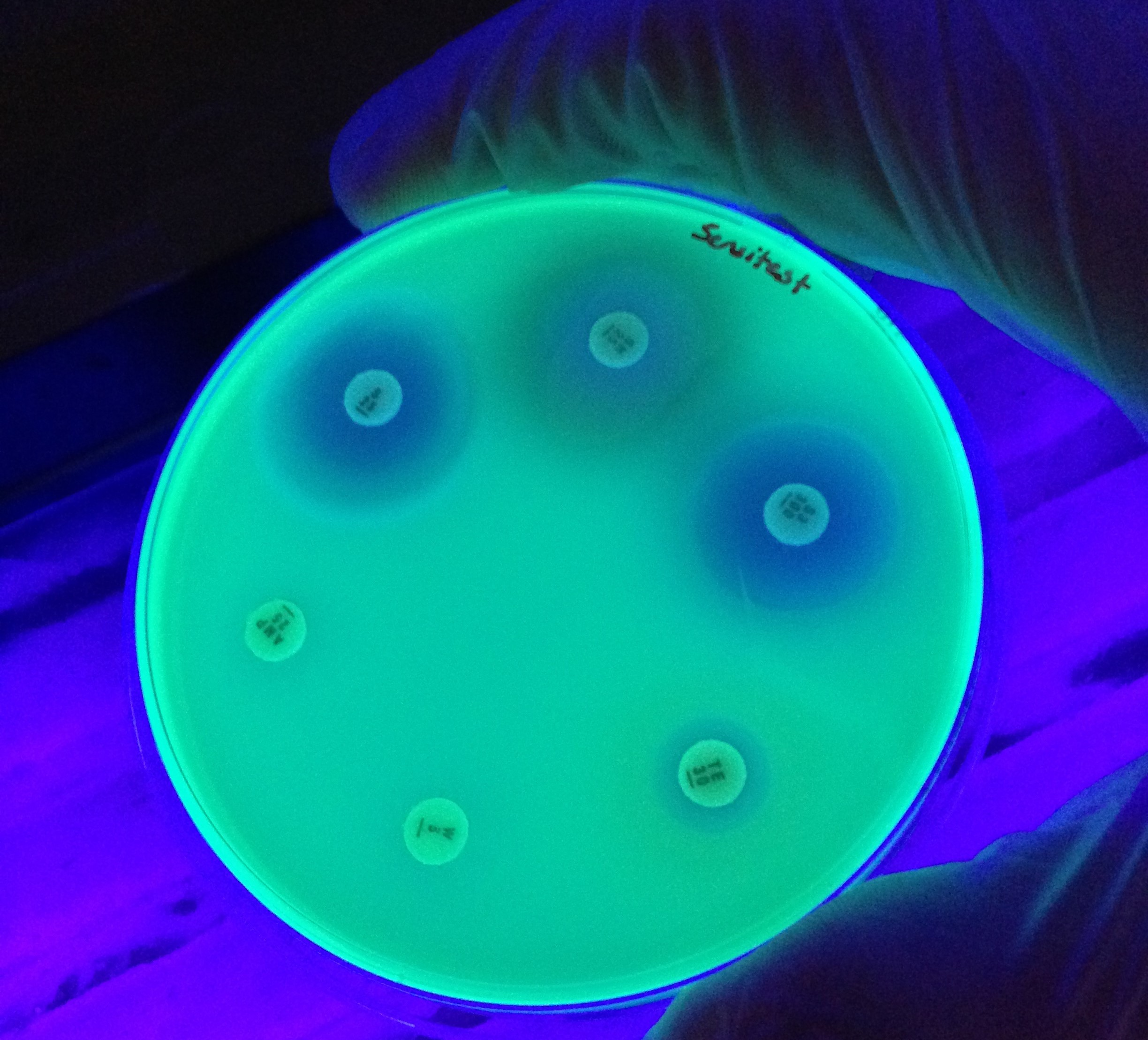
Antimicrobial drugs underpin much of modern medicine. Bacteria, fungi, parasites and viruses that exhibit resistance to antimicrobials threaten the efficacy of therapeutics and impose significant global healthcare and economic burdens. In the midst of the ongoing COVID-19 pandemic, the emergence of pan-drug resistant bacterial and fungal infections in hospitals is a stark reminder of the looming antimicrobial resistance crisis.
That bacteria would develop resistance to antibiotics through mutation was predicted as early as the discovery of penicillin, but the observation of resistance gene transfer between strains in the 1950s heralded the beginning of a period of rapid discovery of resistance mechanisms. Research into the mechanisms of antibiotic resistance proceeded alongside the development of the field of molecular biology and the decades since have seen tremendous advances in our understanding. Still, there is much to learn about where, when and how multi-drug resistance evolves, which organisms are involved and how chains of transmission might be broken before clinicians are faced with intractable infections. Today, multidisciplinary research seeks to answer these questions and the latest advances in sequencing technologies and analysis capacities promise insights into the dynamics of resistance from single cells to complex microbial communities.
Guest-edited by Prof. Willem van Schaik and Dr. Robert Moran, this Antimicrobial Resistance special collection aims to highlight research on the emergence, accumulation and spread of antimicrobial resistance, with a particular focus on opportunistic pathogens and the mobile genetic elements therein.
-
-
-
Arboviruses and their Vectors
More Less
Curated by Journal of General Virology Editor Dr Eng Eong Ooi (Duke NUS Medical School) and Advisory Board Member Dr Esther Schnettler (Bernhard Nocht Institute for Tropical Medicine), this collection presents the latest advances in arbovirus research. This collection was launched in conjuction with IMAV 2017 and in line with IMAV 2019 welcomes submissions of original research articles, Insight Reviews and full-length Reviews.
Find out more about how to submit to the collection here.
-
-
-
Avian Infectious Diseases
More Less
Infectious diseases continue to threaten the sustainability, productivity and growth of the poultry industry worldwide and some present a risk to public health. Many are also present in wild bird populations, with the potential to spill over into domestic birds.
Previously curated by Professor Paul Britton and Dr Mike Skinner, in line with the ‘Pathogenesis and Molecular Biology of Avian Viruses’ Focused Meeting, this collection and the Focused Meeting have been expanded to include bacteria and parasites.
Guest-edited by organisers of the Avian Infectious Diseases 2021 Focused Meeting, Dr Holly Shelton (The Pirbright Institute) and Dr Andrew Broadbent (The Pirbright Institute and University of Maryland), this collection presents high-quality work from Journal of General Virology and Journal of Medical Microbiology on important avian pathogens: their interactions with the host, cell biology, molecular epidemiology and methods of control.This collection is open for submissions in both participating journals, Journal of General Virology and Journal of Medical Microbiology.
Image credit: iStock/Sonja Filitz.
-
-
-
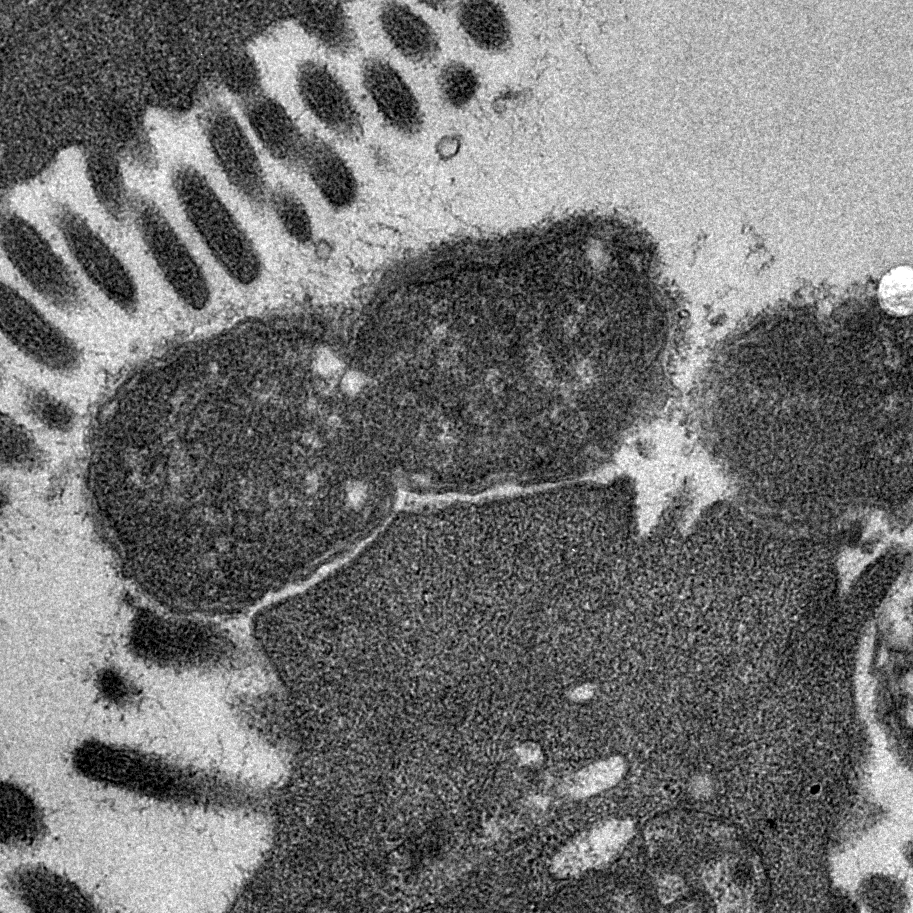
Bacterial Cell Envelopes
More Less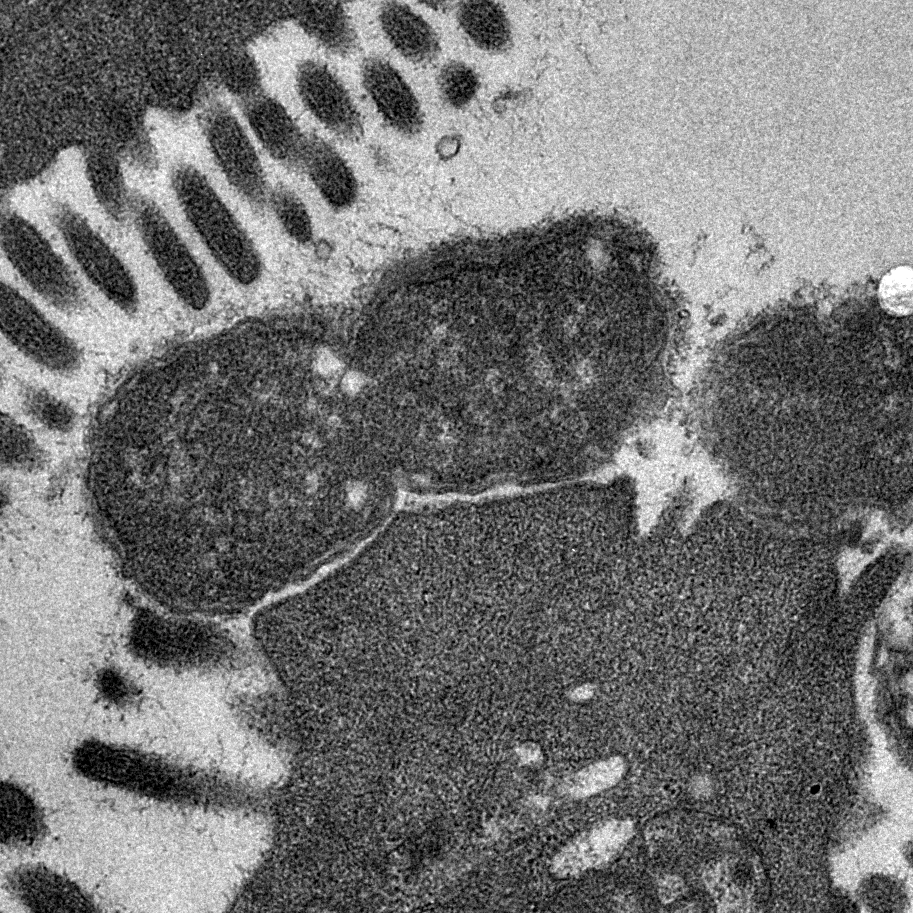 The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
Over the years, Microbiology has published many important findings that have contributed enormously to our understanding of the structure, function and biogenesis of bacterial envelopes. Here we celebrate the journal’s 75th year with a special collection of reviews guest-edited by Professor Tracy Palmer and Dr Yinka Somorin that highlights some of the most important areas of current research in this vibrant research field.
Image shows transmission electron micrograph of a dividing Citrobacter rodentium cell attached to infected mouse colonic epithelium. Characteristic ‘Attaching and Effacing lesions’ or raised pedestals can be seen beneath adhered C. rodentium, formed by the accumulation of host cell actin mediated by the bacterial type III protein secretion system. Effacement of the local brush border villi around the site of attachment is also observed. Courtesy of Dr James Connolly (Newcastle University).
-
-
-
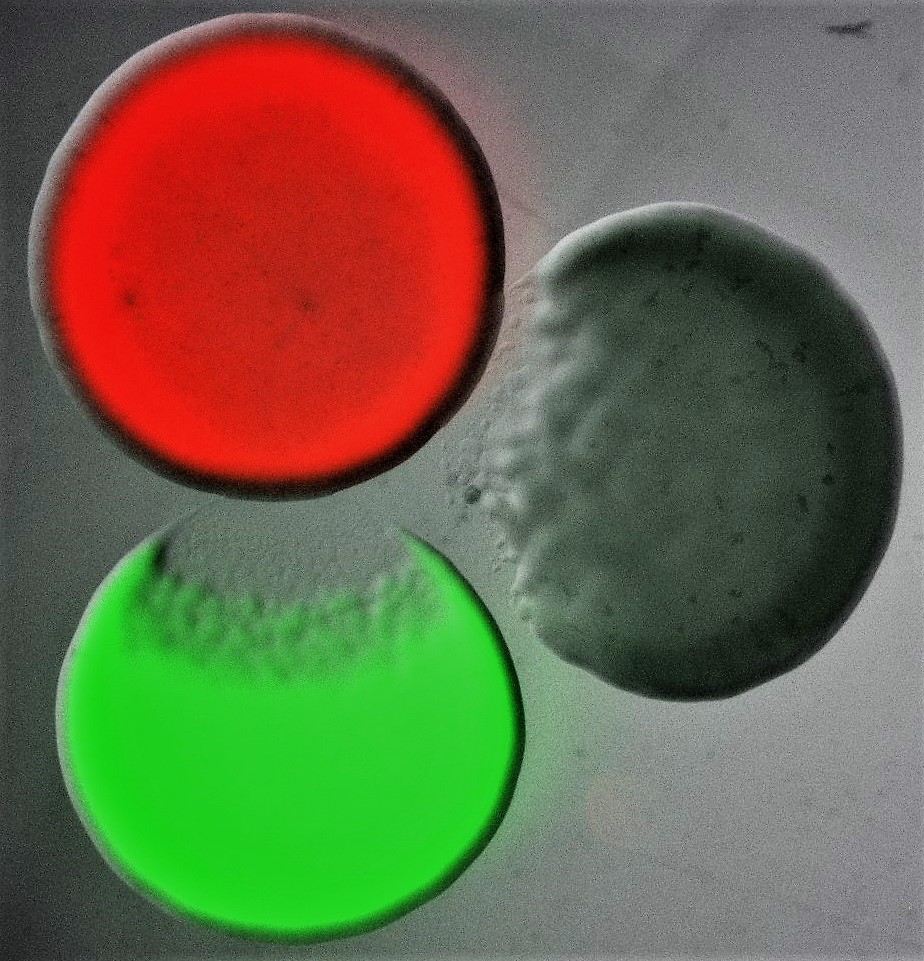
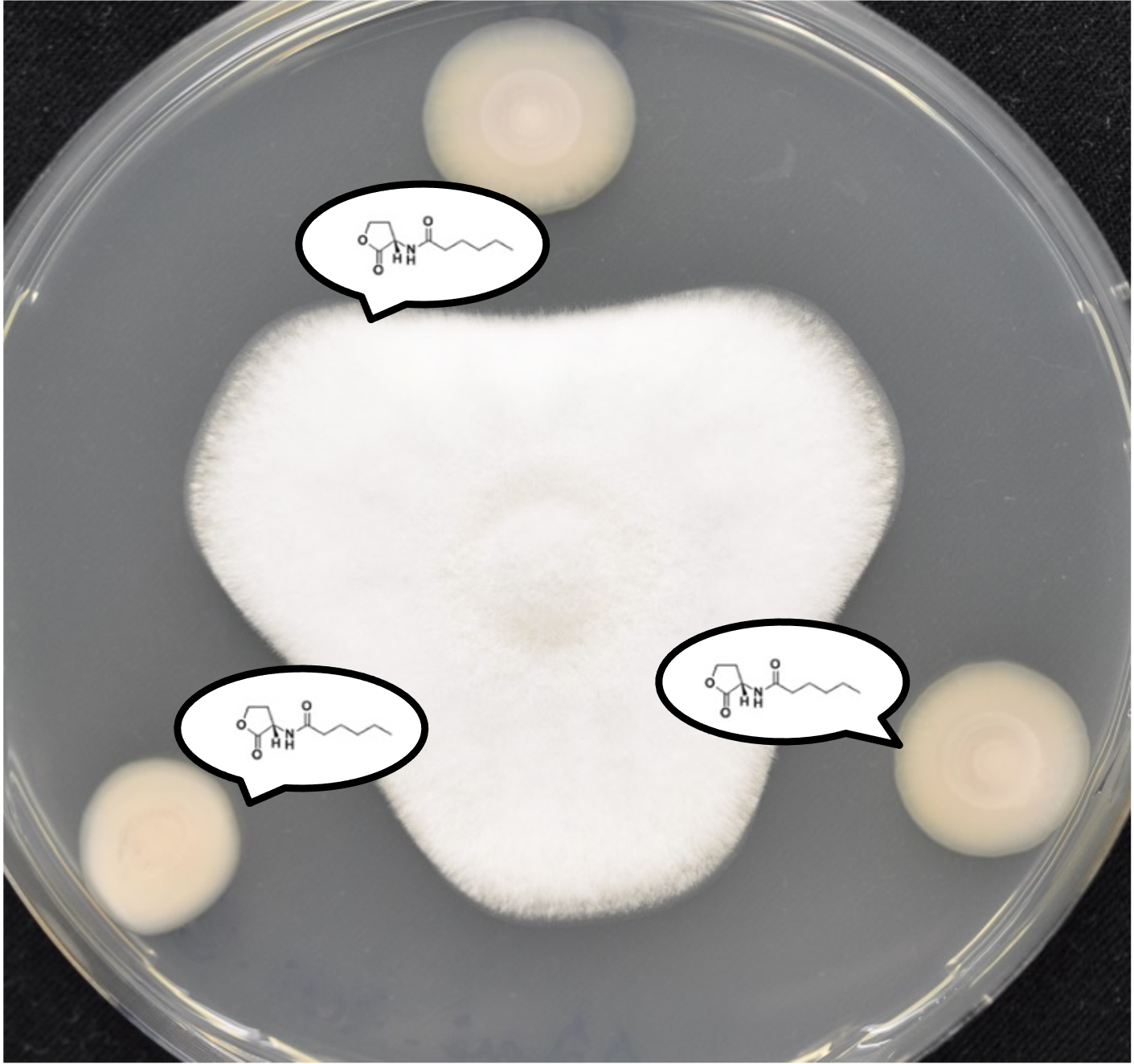
Bacterial Competitive Mechanisms
More Less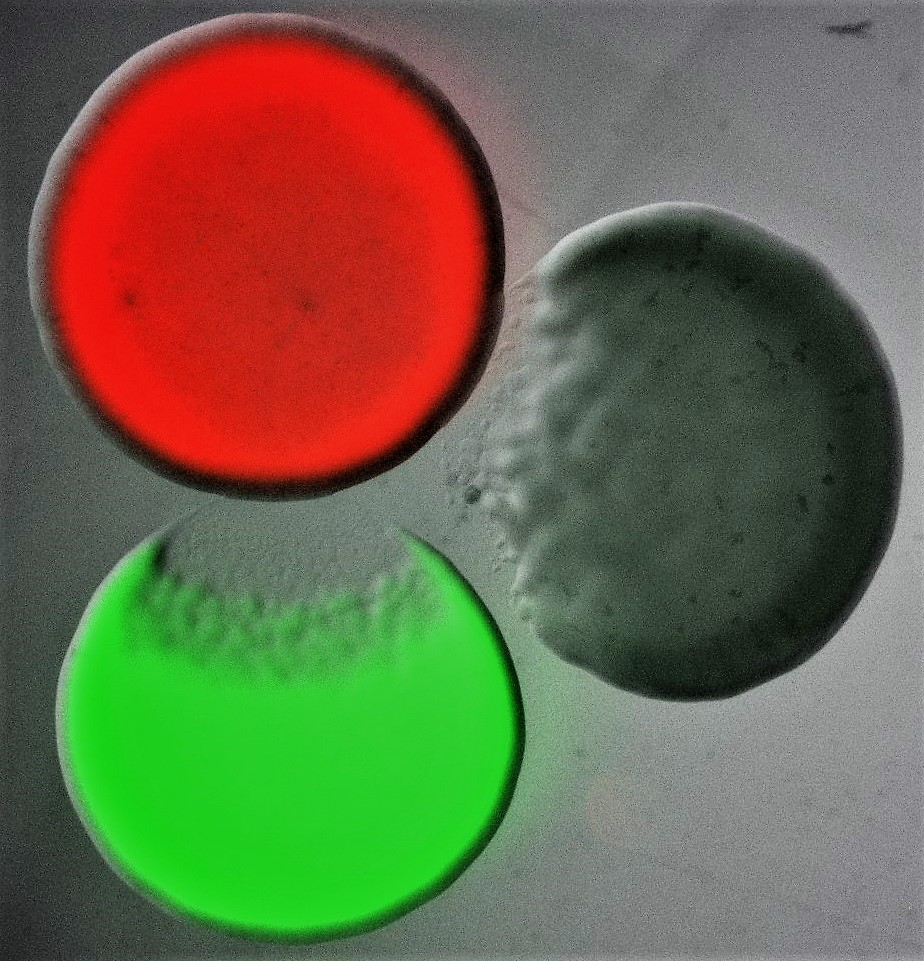
Bacteria are found in every environment and live in complex communities, which are characterized by large densities and diverse compositions. In these microbiomes, bacteria constantly compete for resources, such as nutrients, space and oxygen. Bacteria use a vast arsenal to eliminate their competitors, from elaborate contact-dependent nano-harpoons to small diffusible toxic compounds. In addition to producing these, oftentimes, elaborate weapons, bacteria also employ a range of collective behaviors that, in turn, allow them to carefully regulate weapon production, coordinate their attack strategies with their clonemates, and distinguish friends from foe. In essence, these microscopic warriors take advantage of complex regulation and interbacterial interaction mechanisms to balance the costs of weapon production and deployment with the benefits of gaining precious resources. As such, they manage to not only survive in their highly competitive microcosms, but to also build stable communities that are often crucial for the health of their animal, plant and human hosts.
Over the years, Microbiology has published many studies that have expanded our understanding of the competitive mechanisms bacteria use to thrive in their niches, including, but not limited to, the molecular characterization of toxins or secretion systems and the investigation of interbacterial competition at the organismal level. Here, we take the opportunity to celebrate the journal’s transition to fully Open Access with a special collection of articles that highlights some of the most important areas of current research in this vibrant research field. This collection is guest edited by Professor Despoina A.I. Mavridou (The University of Texas at Austin, USA), Professor Tracy Palmer (Newcastle University, UK) and Dr Sabrina L. Slater (The University of Texas at Austin, USA).
Image shows three differentially labelled Escherichia coli colonies, each of which is producing a different colicin toxin. Colicins are part of the bacteriocin toxin family, an ensemble of diffusible proteinaceous toxins that bacteria release, usually by lysis, to kill phylogenetically similar strains and species. Bacterial killing can be seen characteristically as an area of clearance at the edge of two of the three colonies. Courtesy of Despoina A.I. Mavridou (The University of Texas at Austin).
-
-
-
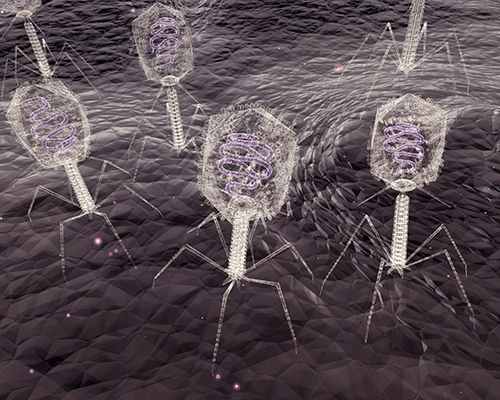
Bacteriophage
More LessBacteriophages were discovered 100 years ago and have since been a key tool used in biological research. Initially used as model organisms for work in genetics and molecular biology, bacteriophages are now known to be one of the major drivers of bacterial evolution and diversification. The introduction of sequencing technologies, phage genomics and metagenomics has highlighted their tremendous diversity and roles in controlling ecological systems within a range of environments. Due to their specificity, phage genomes are now also being manipulated as therapeutics, potentially providing an alternative to conventional antibiotics. Giant phages have also discovered and may represent a novel genus of living organism.Guest-edited by Professor Tetsuya Hayashi (Kyushu University), this collection brings together original Research Articles, Methods, Mini Reviews, and full-length Reviews relating to the diversity of bacteriophages and genomics-based research with a focus on their roles in the evolution of bacteria and ecosystems. Microbial Genomics also welcomes large-scale genomic analyses of bacteriophages which provide the basis for developing novel phage therapies.
This collection is now open for submissions. Submit here and state that your manuscript is part of the Bacteriophage Collection.
-
-
-
Candida
More Less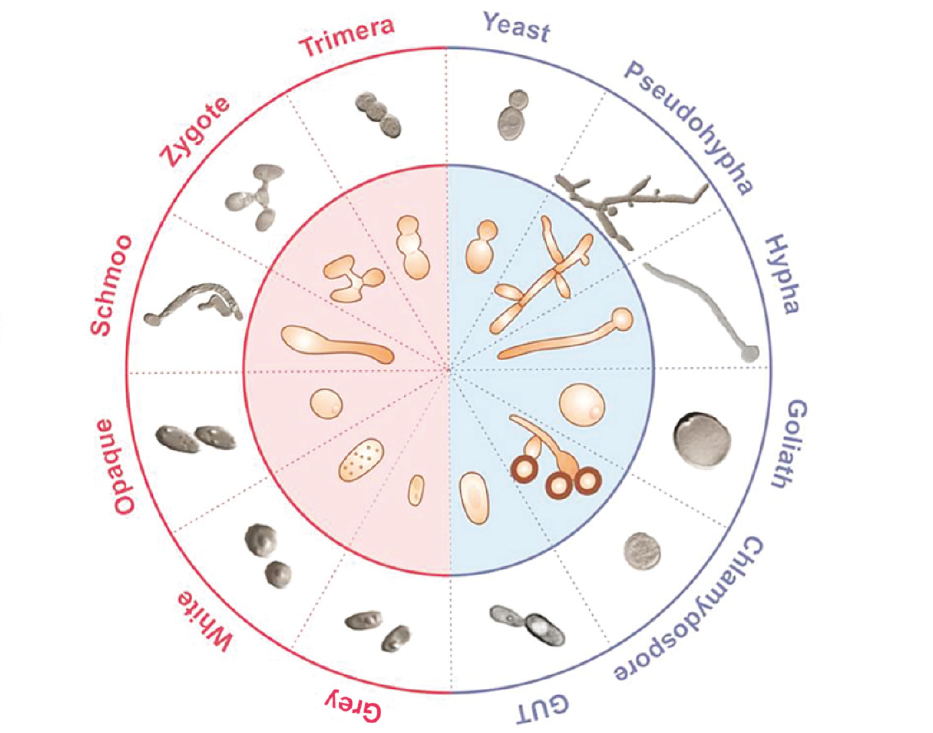
In conjunction with the Candida and Candidiasis meeting, Microbiology Society is bringing together research on Candida biology – species that are major causes of infectious disease in AIDS patients, cancer chemotherapy patients, premature infants, etc. The polymorphic yeast Candida albicans is the most important fungal pathogen in humans. Beyond the clinics, basic research in this organism deals with a variety of topics of interest, like the genetics and molecular biology behind antifungal drug resistance; the molecular determinants of endurance to nutritional, pH and oxidative stress imposed by the host defences; or its interactions with both the host epithelia and bacterial partners that share the mucosal microbiota with the fungus. This collection of research articles published on diverse aspects of Candida biology showcases the journals’ range of Candida research. This collection is open for submissions across our portfolio. Authors are invited to submit on any aspect of Candida research. Upon submission, please indicate that your manuscript is to be considered for the Candida collection.
-
-
-
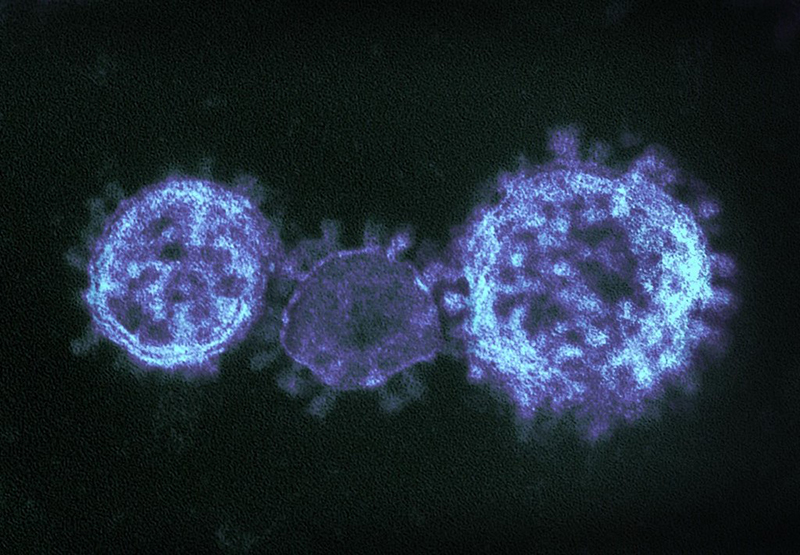
Coronaviruses
More Less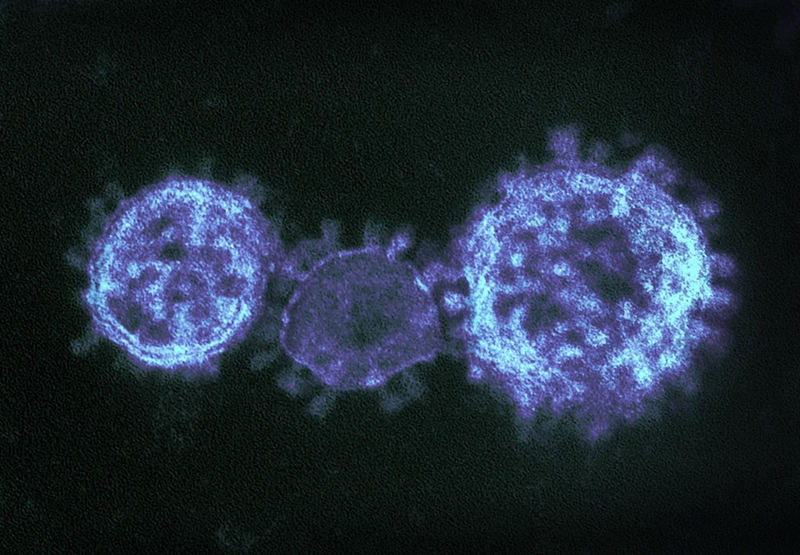
Coronaviruses are a large family of viruses that can infect a range of hosts. They are known to cause diseases including the common cold, Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) in humans.
In January 2020, China saw an outbreak of a new coronavirus strain now named SARS-CoV-2. Although the animal reservoir for the SARS and MERS viruses are known, this has yet to have been confirmed for SARS-CoV-2. All three strains are transmissible between humans.
To allow the widest possible distribution of relevant research, the Microbiology Society has brought together articles from across our portfolio and made this content freely available.
Image credit: "MERS-CoV" by NIAID is licensed under CC BY 2.0, this image has been modified.
-
-
-
CRISPR
More Less
The discovery of the CRISPR-Cas system has led to a sea of change in the microbiological sciences. Since then laboratories all over the world have joined the race to understand and exploit CRISPR. The insights gained about this system led to applications in industry to protect bacterial species against their viral parasites. In addition, CRISPR-Cas has been turned into a versatile genome editing method that has the potential to treat human genetic diseases.
Find out more about CRISPR in this collection of articles by watching this video.
Image credit: iStock/ibreakstock
-
-
-
David Rowlands collection
More Less
Each year, the Microbiology Society Council offer Honorary Membership to distinguished microbiologists who have made a significant contribution to the science. In 2019, David J. Rowlands (Emeritus Professor of Virology, University of Leeds) was appointed an Honorary Member.
This collection brings together Journal of General Virology articles authored by David Rowlands.
-
-
-
Diversity in Microbiology
More Less
As the President of the Microbiology Society, I am eager that we continue to build on our work supporting Equality, Diversity and Inclusion (EDI). Greater diversity within all that we do will widen the talent pool available for the field of microbiology and create networks of ideas and collaborations, potentially leading to greater development and innovation. Working to be inclusive helps us ensure we have a thriving community, which in turn will pave the way for us to support microbiology into the future. I have commissioned a number of articles from under-represented groups, highlighting the talent we have within our Society, and where our members are producing cutting-edge research within our discipline.
Professor Gurdyal Besra, Microbiology Society President
Photo credit: iStock/ Angelina Bambina
-
-
-
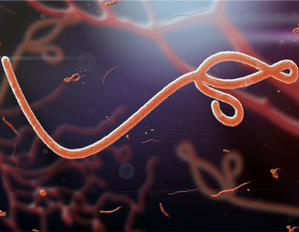
Ebola Virus Disease (EVD)
More Less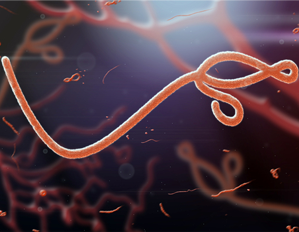
Ebola first became headline news in 2014–2016 when it was transmitted throughout West Africa. In 2019, this severe and often fatal disease has once again been declared a public health emergency of international concern (PHEIC) with over 1700 deaths in this latest outbreak. With vaccines now available, this outbreak could be contained, but only with increased production and delivery of vaccines within the Democratic Republic of Congo.
This collection brings together articles from our portfolio of journals on Ebola virus disease. The Microbiology Society has made this content freely available in the interests of widest possible distribution of relevant research.
-
-
-
Environmental Sensing and Cell-Cell Communication
More Less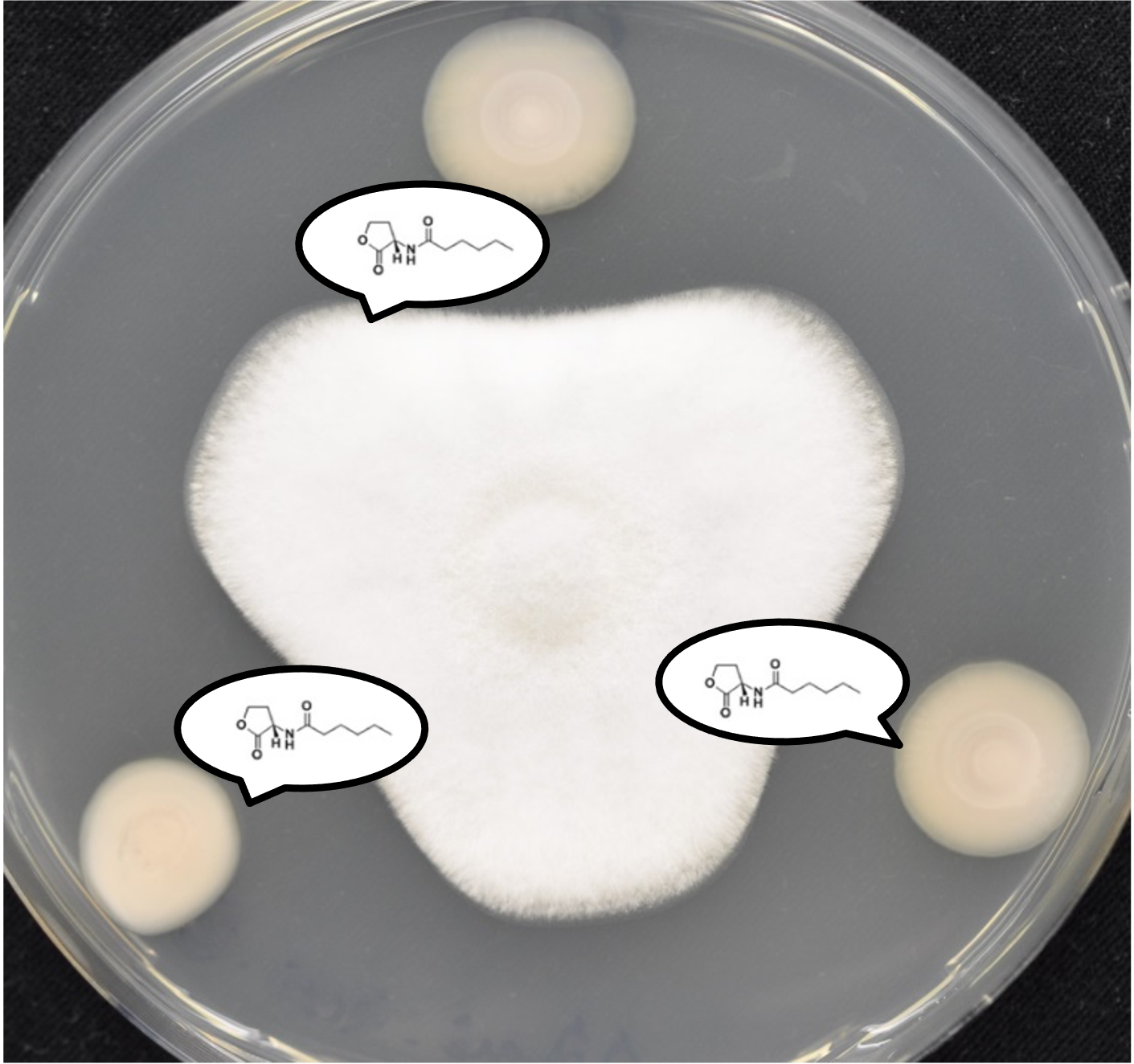
The last two decades have provided a wealth of new insight into how microbes (both prokaryotes and eukaryotes) sense and respond to their surroundings and to one another. Technological advances continue to shape our understanding of this burgeoning field, and this has led to a sea-change in the way in which we view the microbial world. No longer are microbes viewed as being the archetypal single celled entities; instead, community spirit and coordinated responses are the order of the day. In this special anniversary collection for Microbiology, timed to coincide with the Microbiology Society-sponsored Cell-Cell Communication meeting, Guest Editors Martin Welch (University of Cambridge) and Anugraha Mathew (University of Zurich) aim to assemble a landmark collection of papers that celebrate the interaction of microbes with their environment and with one another.
Submissions are particularly welcomed on microbial sensing and signaling pathways, quorum sensing (including both intra- and inter-species interactions and other forms of community-wide behaviors), chemoreception, secondary metabolism, and the complex interplay between different sensory pathways.
-
-
-
Establishing whole genome sequencing at the core of epidemiological surveillance
More Less
Over the last two decades, genome sequencing has become an important tool for understanding and tracking the spread of pathogens. Genomic epidemiology is now a preferred method of surveillance and recent years have seen pathogen sequencing at an unprecedented scale, pushing the underlying technologies to the limit. This has brought major innovations and opportunities to public attention, as well as identifying new research areas. However, major challenges remain in public health settings. These include: incorporating new sequencing technologies and data types for real-time surveillance; developing platforms and nomenclatures for genome-based typing and epidemiology; understanding pathogen evolution and the emergence of virulence and antimicrobial resistance; contextualizing knowledge of clinical microbiology with One Health ecological genomics. In this collection, we bring together recent studies that are establishing pathogen genomics as a major part of contemporary disease control efforts.
-
-
-
Ethnopharmacology
More Less
A concerted effort to characterise, assess and exploit the extensive written and oral record of natural products used in pre-modern European medicine has not been made. This is despite the presence in pre-modern European medical texts of natural products known to be effective in vivo (e.g. Artemisia spp. were used to treat malaria in medieval England) or shown to possess antimicrobial and/or immunomodulatory qualities in vitro (e.g. Allium spp., Plantago spp., Urtica spp.). Given rising antimicrobial resistance and a stalled R&D pipeline for compounds to treat and prevent infection, a thorough scientific evaluation of European ethnopharmacology is overdue.
This collection brings together original articles, mini-reviews, and full-length reviews written by researchers from diverse fields including microbiology, chemistry, botany and the history of medicine, along with industry contacts, to reveal the current state of the art of the field and define areas for collaboration, methods development and translational research.
-
-
-
Exploring the skin microbiome in health and disease
More LessBeing our biggest organ and a primary protective barrier from pathogens, the microbiome of this highly varied environment is now being described in unprecedented detail, revealing complex multispecies communities that play important roles in skin health, the development of the immune system and in wound healing. This collection aims to bring together knowledge of what these microbes are, how they colonise this often nutrient poor and challenging niche, and how they work together to suppress the growth of pathogens, while in themselves also being potential accidental pathogens if the skin barrier is broken.
This collection will feature new primary research and review articles arising from the “Exploring the skin microbiome in health and disease” symposium held at the Microbiology Society Annual Conference 2024 in Edinburgh, 8-11 April 2024.
The collection is also open for new submissions from all researchers across the skin microbiome field. Please indicate within your submission that it is intended for the collection.
Guest Editors: Georgios Efthimious (University of Hull, UK); Albert Bolhois (University of Bath, UK); Andrew Edwards (Imperial College London, UK)
Status: Open for submissions
Deadline for submissions: 7th October 2024
Journals submission links: Microbiology Editorial Manager
Journal of Medical Microbiology Editorial Manager
Microbial Genomics Editorial Manager
-
-
-
Feed the World
More Less
With the United Nations' resolution to eliminate world hunger by 2030 and the globe's population heading towards nine billion, the agriculture industry needs to increase livestock production from the same, or less, land. Livestock uses the most agricultural land (80% including grazing land and cropland for feed). Africa and Asia are the continents with the largest share of the world's uncultivated land, but attempts to develop and expand current capacity in order to meet the growing food demand are halted by deadly killers in the form of viruses, bacterial and protozoan parasites. As such, this area of research is hugely important to ensuring the availability of food for the world’s population.
The ‘Feed the World’ collection brings together articles published across the journal portfolio, focussing on food security, and agriculture and livestock diseases that have an economic impact on humans and animals. Guest edited by Alison Mather (Quadram Institute) and Nigel French (University of New Zealand) for Microbial Genomics; Colin Crump (University of Cambridge) for Journal of General Virology, and Sharon Brookes (Animal Plant Health Agency) for Journal of Medical Microbiology.
This collection is now accepting submissions via any participating journal. Please indicate that your submission is part of the ‘Feed the World’ collection.
-
-
-
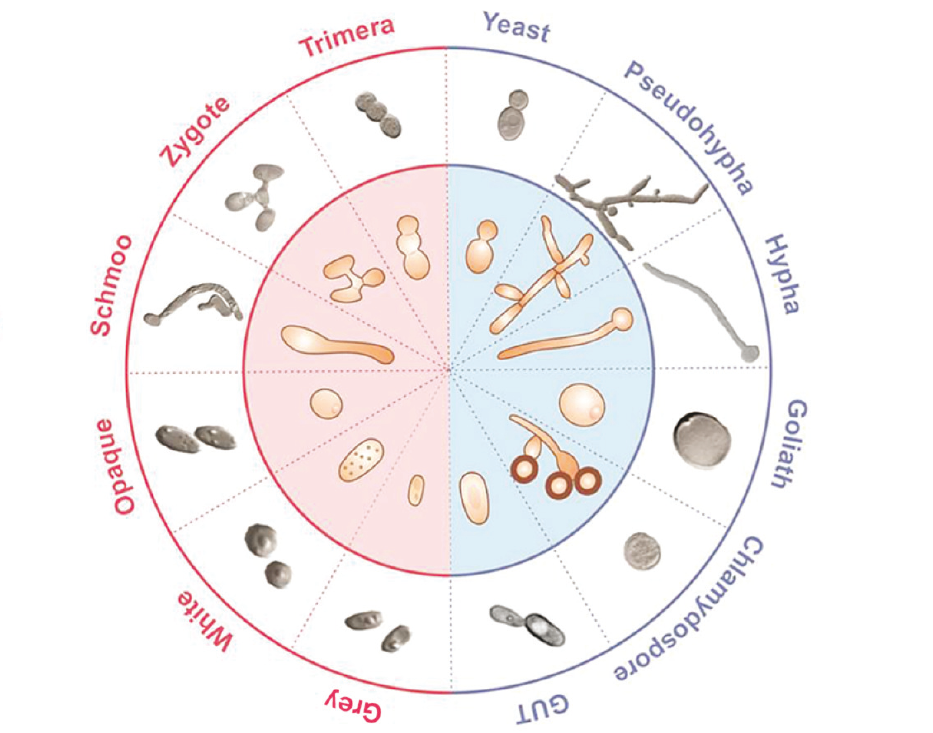
Fungal spotlight: Host-associated microbiomes
More Less
Fungi comprise a distinct eukaryotic lineage often associated with their important role as degraders of organic substrates in the environment. However, fungi also form well-known symbiotic association with higher plants, as mycorrhizae, or with algae as lichens. Additionally, fungi are a critical component of the environmental microbiome associated with both plants and animals. The functional role of fungi in these interactions is poorly understood. Increasingly fungi are being recognized as opportunistic pathogens of animals, including humans, and as plant pathogens, are a threat to the global supply of food. Fungi themselves may harbour their own unique microbiome or organise the microbiome of the substrate they colonise.
This collection will feature studies of fungi in host-associated microbiomes, functional analysis of host-fungal or fungal-microbe interactions, the genetic and genomic diversity of host-associated fungi, as well as the impacts of environmental fungi in natural and manmade ecosystems. It is guest edited by Professor Corby Kistler (University of Minnesota), Dr. Ferry Hagen (Westerdijk Fungal Biodiversity Institute), Dr. David Fitzpatrick (Maynooth University), and Dr. Daniel Croll (University of Neuchâtel).
Image credit: Corby Kistler (University of Minnesota), US Department of Agriculture (public domain), and Biotec, Thailand
-