 -
Volume 165,
Issue 4,
2019
-
Volume 165,
Issue 4,
2019
Volume 165, Issue 4, 2019
- Review
-
-
-
Staphylococcus aureus: setting its sights on the human innate immune system
More LessStaphylococcus aureus has colonized humans for at least 10 000 years, and today inhabits roughly a third of the population. In addition, S. aureus is a major pathogen that is responsible for a significant disease burden, ranging in severity from mild skin and soft-tissue infections to life-threatening endocarditis and necrotizing pneumonia, with treatment often hampered by resistance to commonly available antibiotics. Underpinning its versatility as a pathogen is its ability to evade the innate immune system. S. aureus specifically targets innate immunity to establish and sustain infection, utilizing a large repertoire of virulence factors to do so. Using these factors, S. aureus can resist phagosomal killing, impair complement activity, disrupt cytokine signalling and target phagocytes directly using proteolytic enzymes and cytolytic toxins. Although most of these virulence factors are well characterized, their importance during infection is less clear, as many display species-specific activity against humans or against animal hosts, including cows, horses and chickens. Several staphylococcal virulence factors display species specificity for components of the human innate immune system, with as few as two amino acid changes reducing binding affinity by as much as 100-fold. This represents a major issue for studying their roles during infection, which cannot be examined without the use of humanized infection models. This review summarizes the major factors S. aureus uses to impair the innate immune system, and provides an in-depth look into the host specificity of S. aureus and how this problem is being approached.
-
-
- Microbiology Society Prize Lecture
-
-
-
Transcription activation in bacteria: ancient and modern
More Less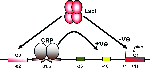 Regulatory interactions at the lac promoter.
Regulatory interactions at the lac promoter.Activation of the transcription of genes is central to many processes of adaptation and differentiation in bacteria. Here, I review the molecular mechanisms by which transcription factors can activate the initiation of specific transcripts at bacterial promoters. The story is presented in the context of Marjory Stephenson’s pioneering work on enzymatic adaptation in bacteria, and sets the different mechanisms in the greater context of how transcription regulatory mechanisms evolved.
-
-
- Biotechnology
-
-
-
Characterization of 4-guanidinobutyrase from Aspergillus niger
More LessArginase is the only fungal ureohydrolase that is well documented in the literature. More recently, a novel route for agmatine catabolism in Aspergillus niger involving another ureohydrolase, 4-guanidinobutyrase (GBase), was reported. We present here a detailed characterization of A. niger GBase – the first fungal (and eukaryotic) enzyme to be studied in detail. A. niger GBase is a homohexamer with a native molecular weight of 336 kDa and an optimal pH of 7.5. Unlike arginase, the Mn2+ enzyme from the same fungus, purified GBase protein is associated with Zn2+ ions. A sensitive fluorescence assay was used to determine its kinetic parameters. GBase acted 25 times more efficiently on 4-guanidinobutyrate (GB) than 3-guanidinopropionic acid (GP). The Km for GB was 2.7±0.4 mM, whereas for GP it was 53.7±0.8 mM. While GB was an efficient nitrogen source, A. niger grew very poorly on GP. Constitutive expression of GBase favoured fungal growth on GP, indicating that GP catabolism is limited by intracellular GBase levels in A. niger. The absence of a specific GPase and the inability of GP to induce GBase expression confine the fungal growth on GP. That GP is a poor substrate for GBase and a very poor nitrogen source for A. niger offers an opportunity to select GBase specificity mutations. Further, it is now possible to compare two distinct ureohydrolases, namely arginase and GBase, from the same organism.
-
-
- Environmental Biology
-
-
-
Potassium resistance of halotolerant and alkaliphilic Halomonas sp. Y2 by a Na+-induced K+ extrusion mechanism
More LessIn most halophiles, K+ generally acts as a major osmotic solute for osmotic adjustment and pH homeostasis. However, strains also need to extrude excessive intracellular K+ to avoid its toxicity. In the halotolerant and alkaliphilic Halomonas sp. Y2, an Na+-induced K+ extrusion process was observed when the cells were confronted with high extracellular K+ pressure and supplementation by millimolar Na+ ions. Among three mechanosensitive channels (KefA) and two K+/H+ antiporters founded in the genome of the strain, ke1 displayed around 3–5-fold upregulation to ion stress at pH 8.0, while much higher upregulation of Ha-mrp was observed at pH 10.0. Compared to the growth of wild-type Halomonas sp. Y2, deletion of these genes from the strain resulted in different growth phenotypes in response to the osmotic pressure of potassium. In combination with the transcriptional response of these genes, we proposed that the KefA channel of Ke1 is the main contributor to the K+-extrusion process under weak alkalinity, while the Mrp system plays critical roles in alleviating K+ contents at high pH. The combination of these strategies allows Halomonas sp. Y2 to grow over a range of extracellular pH and ion concentrations, and thus protect cells under high osmotic stress conditions.
-
-
- Genomics and Systems Biology
-
-
-
Enhanced functionalisation of major facilitator superfamily transporters via fusion of C-terminal protein domains is both extensive and varied in bacteria
More LessThe evolution of gene fusions that result in covalently linked protein domains is widespread in bacteria, where spatially coupling domain functionalities can have functional advantages in vivo. Fusions to integral membrane proteins are less widely studied but could provide routes to enhance membrane function in synthetic biology. We studied the major facilitator superfamily (MFS), as the largest family of transporter proteins in bacteria, to examine the extent and nature of fusions to these proteins. A remarkably diverse variety of fusions are identified and the 8 most abundant examples are described, including additional enzymatic domains and a range of sensory and regulatory domains, many not previously described. Significantly, these fusions are found almost exclusively as C-terminal fusions, revealing that the usually cytoplasmic C-terminal end of MFS protein would the permissive end for engineering synthetic fusions to other cytoplasmic proteins.
-
-
- Host-microbe Interaction
-
-
-
Inactivation of the quorum-sensing transcriptional regulators LasR or RhlR does not suppress the expression of virulence factors and the virulence of Pseudomonas aeruginosa PAO1
More LessPseudomonas aeruginosa is an environmental bacterium but is also an opportunistic pathogen. The aim of this work is to evaluate the contribution of P. aeruginosa LasR and RhlR transcriptional regulators of the quorum-sensing response (QSR) to the production of virulence factors, and to its virulence in a mouse abscess model. The QSR is a complex regulatory network that modulates the expression of several virulence factors, including elastase, pyocyanin and rhamnolipids. LasR, when complexed with the auto-inducer 3-oxo-dodecanoyl lactone (3O-C12-HSL), produced by LasI, is at the top of the QSR regulatory cascade since it activates transcription of some genes encoding virulence factors (such as the gene coding for elastase, lasB) and also transcription of both rhlR and rhlI, encoding the synthase of the auto-inducer butanoyl-homoserine lactone (C4-HSL). In turn RhlR, coupled with C4-HSL, activates the transcription of genes encoding for the enzymes involved in pyocyanin and rhamnolipid production. Several efforts have been made to obtain inhibitors of LasR activity that would suppress the QSR. However, these attempts have used chemical compounds that might not be specific for LasR inactivation. In this work we show that individual inactivation of either lasR or rhlR did not block the QSR, nor did it impair P. aeruginosa virulence, and that even a lasR rhlR double mutant still presented residual virulence, even lacking the production of virulence factors. These results show that the inhibition of either lasR or rhlR is not a straightforward approach to blocking P. aeruginosa virulence, due to the great complexity of the QSR.
-
-
- Physiology and Metabolism
-
-
-
Apocarotenoids produced from β-carotene by dioxygenases from Mucor circinelloides
More LessMucor circinelloides exhibits the complex sexual behaviour that is induced in other Mucoromycotina by a family of apocarotenoids called trisporoids. The genome of M. circinelloides contains four genes encoding putative carotenoid cleavage dioxygenases. The gene products of two of them were sufficient to convert β-carotene into the precursors of three families of apocarotenoids, both in vitro and in the Escherichia coli heterologous in vivo system. The first of these products, CarS, cleaved the C40 β-carotene into the C15 precursor of cyclofarnesoids and a C25 apocarotenal that was converted by the second enzyme, AcaA, into the C18 precursor of trisporoids and the C7 precursor of methylhexanoids. Apocarotenoids were not found in single or mixed cultures of the two strains of opposite sex, whose interaction readily produced zygospores, the sexual fusion cells.
-
-
-
-
Very rapid flow cytometric assessment of antimicrobial susceptibility during the apparent lag phase of microbial (re)growth
More Less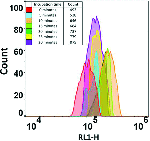 Rapid changes in the number and flow cytometric behaviour of cells of
E. coli
taken from a stationary phase and inoculated into rich medium.
Rapid changes in the number and flow cytometric behaviour of cells of
E. coli
taken from a stationary phase and inoculated into rich medium.Cells of E. coli were grown in LB medium, taken from a stationary phase of 2–4 h, and re-inoculated into fresh media at a concentration (105 ml−1 or lower) characteristic of bacteriuria. Flow cytometry was used to assess how quickly we could detect changes in cell size, number, membrane energization (using a carbocyanine dye) and DNA distribution. It transpired that while the lag phase observable macroscopically via bulk OD measurements could be as long as 4 h, the true lag phase could be less than 15–20 min, and was accompanied by many observable biochemical changes. Antibiotics to which the cells were sensitive affected these changes within 20 min of re-inoculation, providing the possibility of a very rapid antibiotic susceptibility test on a timescale compatible with a visit to a GP clinic. The strategy was applied successfully to genuine potential urinary tract infection (UTI) samples taken from a doctor’s surgery. The methods developed could prove of considerable value in ensuring the correct prescription and thereby lowering the spread of antimicrobial resistance.
-
-
-
Identification and biosynthesis of 2-(1H-imidazol-5-yl) ethan-1-ol (histaminol) in methanogenic archaea
More LessHistaminol is a relatively rare metabolite most commonly resulting from histidine metabolism. Here we describe histaminol production and secretion into the culture broth by the methanogen Methanococcus maripaludis S2 as well as a number of other methanogens. This work is the first identification of this compound as a natural product in methanogens. Its biosynthesis from histidine was confirmed by the incorporation of 2H3-histidine into histaminol by growing cells of M. maripaludis S2. Possible functions of this molecule could be cell signaling as observed with histamine in eukaryotes or uptake of metal ions.
-
- Regulation
-
-
-
Direct inhibition of transcription in vitro by the isolated N-terminal domain of the Escherichia coli nucleoid-associated protein H-NS and by its paralogue Hha
More LessH-NS is an abundant nucleoid-associated protein in the enterobacteria that mediates both chromatin compaction and transcriptional silencing of numerous genes, especially those that have been acquired by horizontal transfer or that are involved in virulence functions. With two dimerization domains (N-terminal and central) and a C-terminal DNA-binding domain, the 15 kDa H-NS polypeptide can assemble as long polymeric filaments on DNA, and mutations in any of the three domains confer a dominant-negative phenotype in vivo by a subunit-poisoning mechanism. Here we confirm that several of these mutants [L26P, I119T and a truncation beyond residue 92(Δ93)] are also dominant-negative in vitro, in that they reverse the inhibition imposed by native H-NS in two different transcription assay formats (initiation+elongation, or elongation alone). On the other hand, another dominant-negative truncation mutant Δ64 (which possesses only the protein's N-terminal domain) per se completely and unexpectedly inhibited transcription in both assay formats. The Hha protein, which is a paralogue of H-NS and resembles its isolated N-terminal domain, also behaved like Δ64 as an inhibitor of transcription in vitro. We propose that under certain growth conditions, Escherichia coli RNA polymerase may be the direct inhibitory target of Hha, and that this effect is experimentally mimicked by the isolated N-terminal domain of H-NS.
-
-
-
-
Functional characterization of BcrR: a one-component transmembrane signal transduction system for bacitracin resistance
More LessBacitracin is a cell wall targeting antimicrobial with clinical and agricultural applications. With the growing mismatch between antimicrobial resistance and development, it is essential we understand the molecular mechanisms of resistance in order to prioritize and generate new effective antimicrobials. BcrR is a unique membrane-bound one-component system that regulates high-level bacitracin resistance in Enterococcus faecalis . In the presence of bacitracin, BcrR activates transcription of the bcrABD operon conferring resistance through a putative ATP-binding cassette (ABC) transporter (BcrAB). BcrR has three putative functional domains, an N-terminal helix–turn–helix DNA-binding domain, an intermediate oligomerization domain and a C-terminal transmembrane domain. However, the molecular mechanisms of signal transduction remain unknown. Random mutagenesis of bcrR was performed to generate loss- and gain-of-function mutants using transcriptional reporters fused to the target promoter P bcrA . Fifteen unique mutants were isolated across all three proposed functional domains, comprising 14 loss-of-function and one gain-of-function mutant. The gain-of-function variant (G64D) mapped to the putative dimerization domain of BcrR, and functional analyses indicated that the G64D mutant constitutively expresses the P bcrA-luxABCDE reporter. DNA-binding and membrane insertion were not affected in the five mutants chosen for further characterization. Homology modelling revealed putative roles for two key residues (R11 and S33) in BcrR activation. Here we present a new model of BcrR activation and signal transduction, providing valuable insight into the functional characterization of membrane-bound one-component systems and how they can coordinate critical bacterial responses, such as antimicrobial resistance.
-
Volumes and issues
-
Volume 170 (2024)
-
Volume 169 (2023)
-
Volume 168 (2022)
-
Volume 167 (2021)
-
Volume 166 (2020)
-
Volume 165 (2019)
-
Volume 164 (2018)
-
Volume 163 (2017)
-
Volume 162 (2016)
-
Volume 161 (2015)
-
Volume 160 (2014)
-
Volume 159 (2013)
-
Volume 158 (2012)
-
Volume 157 (2011)
-
Volume 156 (2010)
-
Volume 155 (2009)
-
Volume 154 (2008)
-
Volume 153 (2007)
-
Volume 152 (2006)
-
Volume 151 (2005)
-
Volume 150 (2004)
-
Volume 149 (2003)
-
Volume 148 (2002)
-
Volume 147 (2001)
-
Volume 146 (2000)
-
Volume 145 (1999)
-
Volume 144 (1998)
-
Volume 143 (1997)
-
Volume 142 (1996)
-
Volume 141 (1995)
-
Volume 140 (1994)
-
Volume 139 (1993)
-
Volume 138 (1992)
-
Volume 137 (1991)
-
Volume 136 (1990)
-
Volume 135 (1989)
-
Volume 134 (1988)
-
Volume 133 (1987)
-
Volume 132 (1986)
-
Volume 131 (1985)
-
Volume 130 (1984)
-
Volume 129 (1983)
-
Volume 128 (1982)
-
Volume 127 (1981)
-
Volume 126 (1981)
-
Volume 125 (1981)
-
Volume 124 (1981)
-
Volume 123 (1981)
-
Volume 122 (1981)
-
Volume 121 (1980)
-
Volume 120 (1980)
-
Volume 119 (1980)
-
Volume 118 (1980)
-
Volume 117 (1980)
-
Volume 116 (1980)
-
Volume 115 (1979)
-
Volume 114 (1979)
-
Volume 113 (1979)
-
Volume 112 (1979)
-
Volume 111 (1979)
-
Volume 110 (1979)
-
Volume 109 (1978)
-
Volume 108 (1978)
-
Volume 107 (1978)
-
Volume 106 (1978)
-
Volume 105 (1978)
-
Volume 104 (1978)
-
Volume 103 (1977)
-
Volume 102 (1977)
-
Volume 101 (1977)
-
Volume 100 (1977)
-
Volume 99 (1977)
-
Volume 98 (1977)
-
Volume 97 (1976)
-
Volume 96 (1976)
-
Volume 95 (1976)
-
Volume 94 (1976)
-
Volume 93 (1976)
-
Volume 92 (1976)
-
Volume 91 (1975)
-
Volume 90 (1975)
-
Volume 89 (1975)
-
Volume 88 (1975)
-
Volume 87 (1975)
-
Volume 86 (1975)
-
Volume 85 (1974)
-
Volume 84 (1974)
-
Volume 83 (1974)
-
Volume 82 (1974)
-
Volume 81 (1974)
-
Volume 80 (1974)
-
Volume 79 (1973)
-
Volume 78 (1973)
-
Volume 77 (1973)
-
Volume 76 (1973)
-
Volume 75 (1973)
-
Volume 74 (1973)
-
Volume 73 (1972)
-
Volume 72 (1972)
-
Volume 71 (1972)
-
Volume 70 (1972)
-
Volume 69 (1971)
-
Volume 68 (1971)
-
Volume 67 (1971)
-
Volume 66 (1971)
-
Volume 65 (1971)
-
Volume 64 (1970)
-
Volume 63 (1970)
-
Volume 62 (1970)
-
Volume 61 (1970)
-
Volume 60 (1970)
-
Volume 59 (1969)
-
Volume 58 (1969)
-
Volume 57 (1969)
-
Volume 56 (1969)
-
Volume 55 (1969)
-
Volume 54 (1968)
-
Volume 53 (1968)
-
Volume 52 (1968)
-
Volume 51 (1968)
-
Volume 50 (1968)
-
Volume 49 (1967)
-
Volume 48 (1967)
-
Volume 47 (1967)
-
Volume 46 (1967)
-
Volume 45 (1966)
-
Volume 44 (1966)
-
Volume 43 (1966)
-
Volume 42 (1966)
-
Volume 41 (1965)
-
Volume 40 (1965)
-
Volume 39 (1965)
-
Volume 38 (1965)
-
Volume 37 (1964)
-
Volume 36 (1964)
-
Volume 35 (1964)
-
Volume 34 (1964)
-
Volume 33 (1963)
-
Volume 32 (1963)
-
Volume 31 (1963)
-
Volume 30 (1963)
-
Volume 29 (1962)
-
Volume 28 (1962)
-
Volume 27 (1962)
-
Volume 26 (1961)
-
Volume 25 (1961)
-
Volume 24 (1961)
-
Volume 23 (1960)
-
Volume 22 (1960)
-
Volume 21 (1959)
-
Volume 20 (1959)
-
Volume 19 (1958)
-
Volume 18 (1958)
-
Volume 17 (1957)
-
Volume 16 (1957)
-
Volume 15 (1956)
-
Volume 14 (1956)
-
Volume 13 (1955)
-
Volume 12 (1955)
-
Volume 11 (1954)
-
Volume 10 (1954)
-
Volume 9 (1953)
-
Volume 8 (1953)
-
Volume 7 (1952)
-
Volume 6 (1952)
-
Volume 5 (1951)
-
Volume 4 (1950)
-
Volume 3 (1949)
-
Volume 2 (1948)
-
Volume 1 (1947)
Most Read This Month


