
Full text loading...
A step-by-step method for creating a tailored 3D culture medium for study of soil microbes. The complete workflow can be split into three parts: growth and observation, metabolic profiling to provide a stable refractive index matching solution, and production of the 3D soil environment. The 3D culture scaffold was created by cryomilling Nafion resin pellets and size filtration. Chemical processing altered the surface chemistry of Nafion particles and facilitated nutrient binding by titration of a defined liquid culture medium. Metabolic profiling determined non-metabolisable sugars and provided an inert refractive index matching substrate, which was added to the final nutrient titration. Inoculation and growth of the test strain allowed for downstream assessment of colonisation behaviours and community dynamics in situ by, for example, optical microscopy.
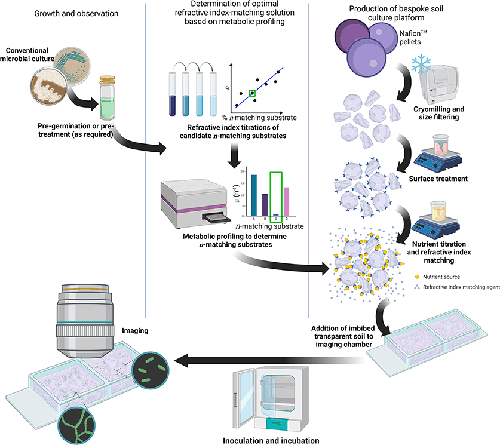
We have developed a tuneable workflow for the study of soil microbes in an imitative 3D soil environment that is compatible with routine and advanced optical imaging, is chemically customisable, and is reliably refractive index matched based on the carbon catabolism of the study organism. We demonstrate our transparent soil pipeline with two representative soil organisms, Bacillus subtilis and Streptomyces coelicolor , and visualise their colonisation behaviours using fluorescence microscopy and mesoscopy. This spatially structured, 3D approach to microbial culture has the potential to further study the behaviour of bacteria in conditions matching their native environment and could be expanded to study microbial interactions, such as competition and warfare.