
Full text loading...
 , Suman Basu1, Mousumi Bhattacharyya1 and Tapan K. Dutta1
, Suman Basu1, Mousumi Bhattacharyya1 and Tapan K. Dutta1
Biochemical and molecular analyses to reveal the regulation of catabolic pathways in the complete degradation of dioctyl phthalate isomers in
Gordonia
sp.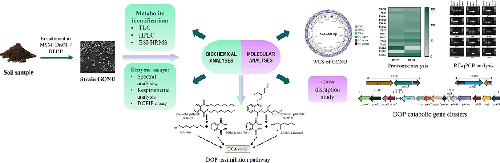
Bacterial strain GONU, belonging to the genus Gordonia , was isolated from a municipal waste-contaminated soil sample and was capable of utilizing an array of endocrine-disrupting phthalate diesters, including di-n-octyl phthalate (DnOP) and its isomer di(2-ethylhexyl) phthalate (DEHP), as the sole carbon and energy sources. The biochemical pathways of the degradation of DnOP and DEHP were evaluated in strain GONU by using a combination of various chromatographic, spectrometric and enzymatic analyses. Further, the upregulation of three different esterases (estG2, estG3 and estG5), a phthalic acid (PA)-metabolizing pht operon and a protocatechuic acid (PCA)-metabolizing pca operon were revealed based on de novo whole genome sequence information and substrate-induced protein profiling by LC-ESI-MS/MS analysis followed by differential gene expression by real-time PCR. Subsequently, functional characterization of the differentially upregulated esterases on the inducible hydrolytic metabolism of DnOP and DEHP revealed that EstG5 is involved in the hydrolysis of DnOP to PA, whereas EstG2 and EstG3 are involved in the metabolism of DEHP to PA. Finally, gene knockout experiments further validated the role of EstG2 and EstG5, and the present study deciphered the inducible regulation of the specific genes and operons in the assimilation of DOP isomers.

Article metrics loading...

Full text loading...
References


Data & Media loading...
Supplements
