Collections
Browse our collections – bringing together peer-reviewed content from across the Society’s publishing platform on a range of hot topics and subject areas.
-
-
Bacterial Cell Envelopes
More Less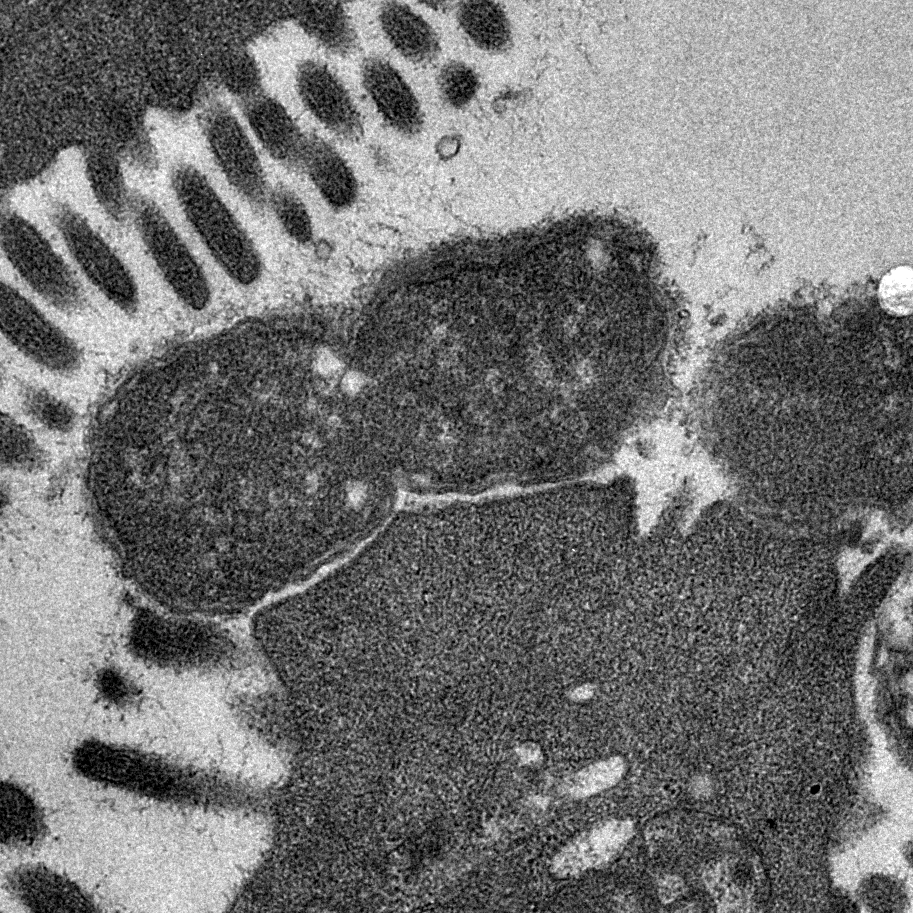 The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
Over the years, Microbiology has published many important findings that have contributed enormously to our understanding of the structure, function and biogenesis of bacterial envelopes. Here we celebrate the journal’s 75th year with a special collection of reviews guest-edited by Professor Tracy Palmer and Dr Yinka Somorin that highlights some of the most important areas of current research in this vibrant research field.
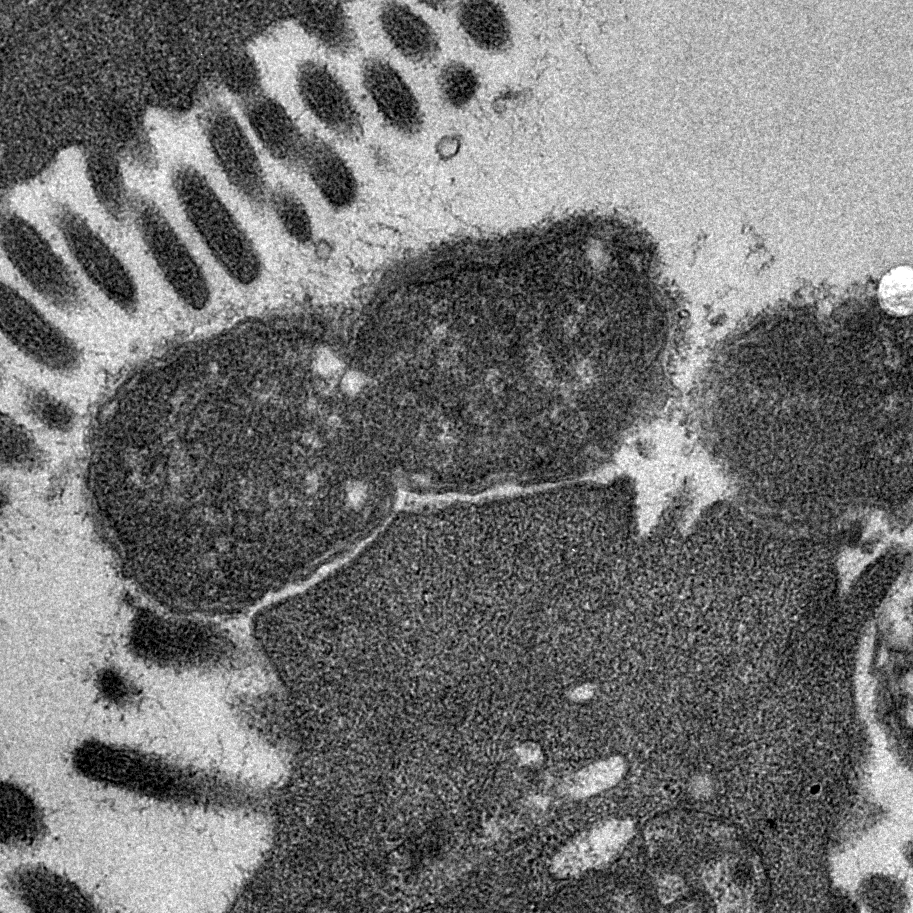
Image shows transmission electron micrograph of a dividing Citrobacter rodentium cell attached to infected mouse colonic epithelium. Characteristic ‘Attaching and Effacing lesions’ or raised pedestals can be seen beneath adhered C. rodentium, formed by the accumulation of host cell actin mediated by the bacterial type III protein secretion system. Effacement of the local brush border villi around the site of attachment is also observed. Courtesy of Dr James Connolly (Newcastle University).
-
-
-
Bacterial Competitive Mechanisms
More Less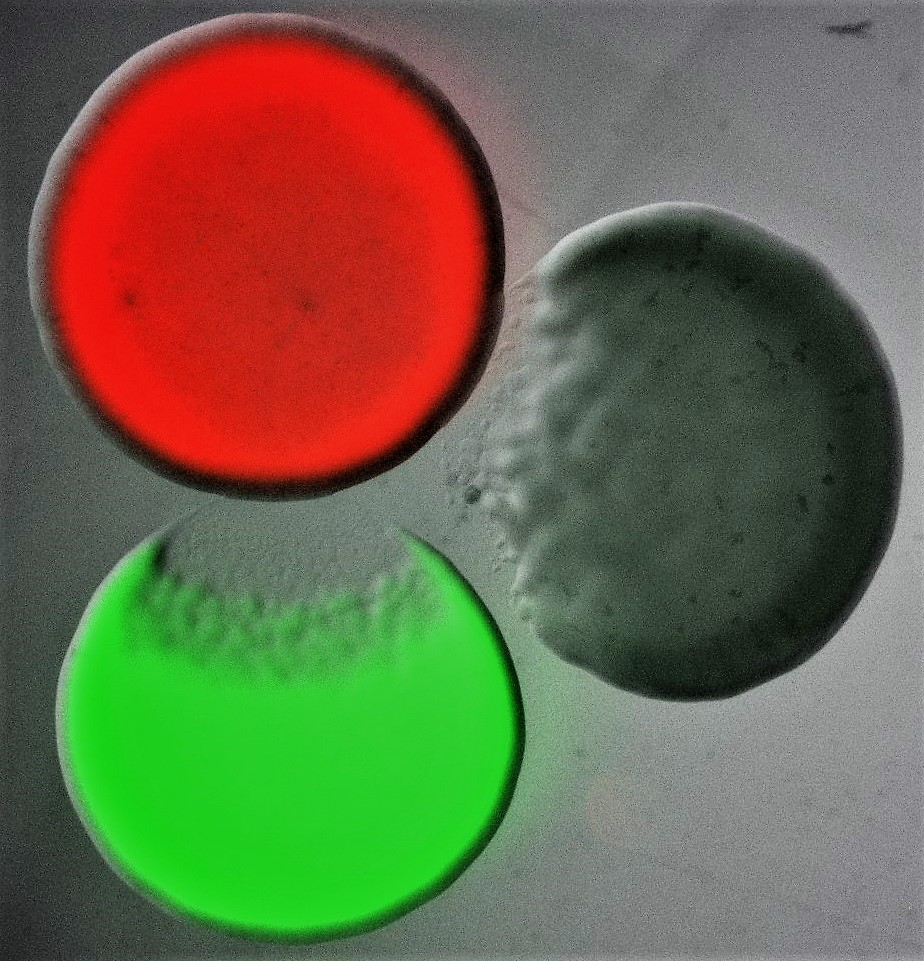
Bacteria are found in every environment and live in complex communities, which are characterized by large densities and diverse compositions. In these microbiomes, bacteria constantly compete for resources, such as nutrients, space and oxygen. Bacteria use a vast arsenal to eliminate their competitors, from elaborate contact-dependent nano-harpoons to small diffusible toxic compounds. In addition to producing these, oftentimes, elaborate weapons, bacteria also employ a range of collective behaviors that, in turn, allow them to carefully regulate weapon production, coordinate their attack strategies with their clonemates, and distinguish friends from foe. In essence, these microscopic warriors take advantage of complex regulation and interbacterial interaction mechanisms to balance the costs of weapon production and deployment with the benefits of gaining precious resources. As such, they manage to not only survive in their highly competitive microcosms, but to also build stable communities that are often crucial for the health of their animal, plant and human hosts.
Over the years, Microbiology has published many studies that have expanded our understanding of the competitive mechanisms bacteria use to thrive in their niches, including, but not limited to, the molecular characterization of toxins or secretion systems and the investigation of interbacterial competition at the organismal level. Here, we take the opportunity to celebrate the journal’s transition to fully Open Access with a special collection of articles that highlights some of the most important areas of current research in this vibrant research field. This collection is guest edited by Professor Despoina A.I. Mavridou (The University of Texas at Austin, USA), Professor Tracy Palmer (Newcastle University, UK) and Dr Sabrina L. Slater (The University of Texas at Austin, USA).
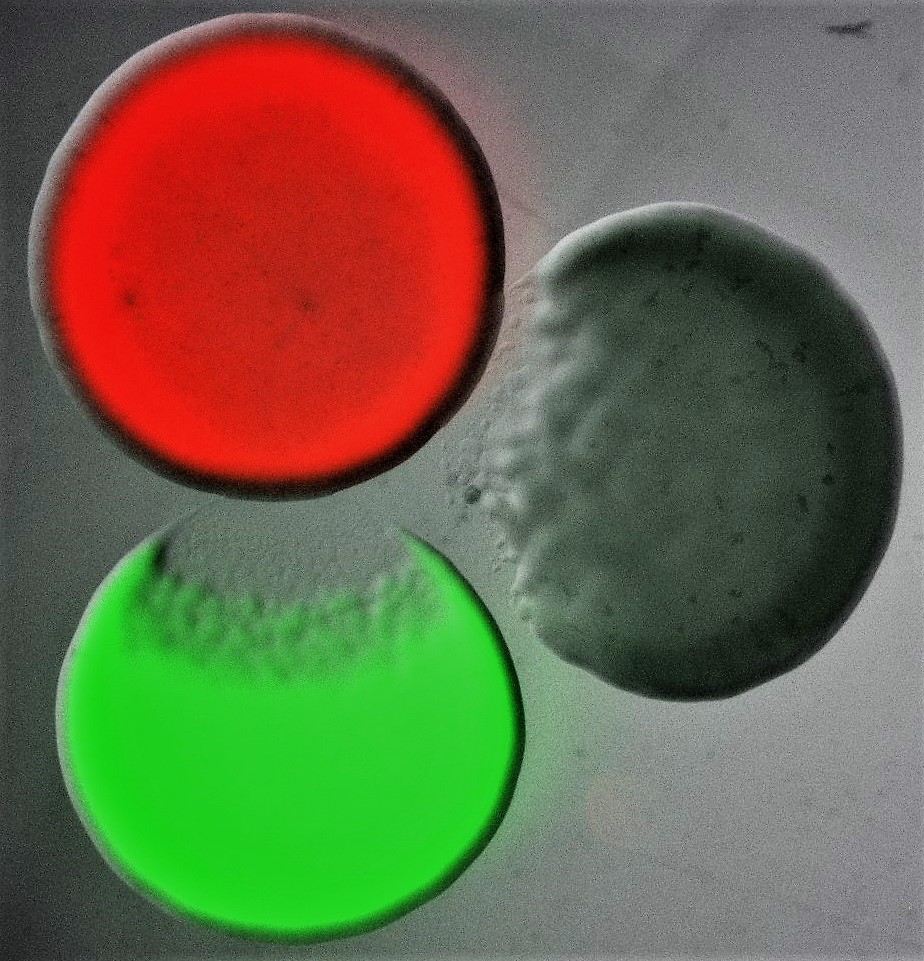
Image shows three differentially labelled Escherichia coli colonies, each of which is producing a different colicin toxin. Colicins are part of the bacteriocin toxin family, an ensemble of diffusible proteinaceous toxins that bacteria release, usually by lysis, to kill phylogenetically similar strains and species. Bacterial killing can be seen characteristically as an area of clearance at the edge of two of the three colonies. Courtesy of Despoina A.I. Mavridou (The University of Texas at Austin).
-
-
-
Bacteriophage
More LessBacteriophages were discovered 100 years ago and have since been a key tool used in biological research. Initially used as model organisms for work in genetics and molecular biology, bacteriophages are now known to be one of the major drivers of bacterial evolution and diversification. The introduction of sequencing technologies, phage genomics and metagenomics has highlighted their tremendous diversity and roles in controlling ecological systems within a range of environments. Due to their specificity, phage genomes are now also being manipulated as therapeutics, potentially providing an alternative to conventional antibiotics. Giant phages have also discovered and may represent a novel genus of living organism.Guest-edited by Professor Tetsuya Hayashi (Kyushu University), this collection brings together original Research Articles, Methods, Mini Reviews, and full-length Reviews relating to the diversity of bacteriophages and genomics-based research with a focus on their roles in the evolution of bacteria and ecosystems. Microbial Genomics also welcomes large-scale genomic analyses of bacteriophages which provide the basis for developing novel phage therapies.
This collection is now open for submissions. Submit here and state that your manuscript is part of the Bacteriophage Collection.
-