Coronaviruses
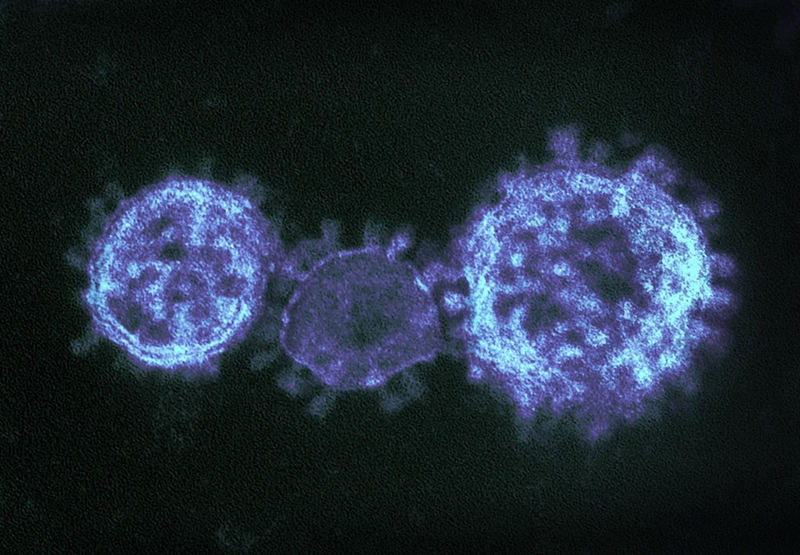
Coronaviruses are a large family of viruses that can infect a range of hosts. They are known to cause diseases including the common cold, Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) in humans.
In January 2020, China saw an outbreak of a new coronavirus strain now named SARS-CoV-2. Although the animal reservoir for the SARS and MERS viruses are known, this has yet to have been confirmed for SARS-CoV-2. All three strains are transmissible between humans.
To allow the widest possible distribution of relevant research, the Microbiology Society has brought together articles from across our portfolio and made this content freely available.
Image credit: "MERS-CoV" by NIAID is licensed under CC BY 2.0, this image has been modified.
Collection Contents
1 - 20 of 298 results
-
-
Evolution of the coronavirus spike protein in the full-length genome and defective viral genome under diverse selection pressures
More LessHow coronaviruses evolve by altering the structures of their full-length genome and defective viral genome (DVG) under dynamic selection pressures has not been studied. In this study, we aimed to experimentally identify the dynamic evolutionary patterns of the S protein sequence in the full-length genome and DVG under diverse selection pressures, including persistence, innate immunity and antiviral drugs. The evolutionary features of the S protein sequence in the full-length genome and in the DVG under diverse selection pressures are as follows: (i) the number of nucleotide (nt) mutations does not necessarily increase with the number of selection pressures; (ii) certain types of selection pressure(s) can lead to specific nt mutations; (iii) the mutated nt sequence can be reverted to the wild-type nt sequence under the certain type of selection pressure(s); (iv) the DVG can also undergo mutations and evolve independently of the full-length genome; and (v) DVG species are regulated during evolution under diverse selection pressures. The various evolutionary patterns of the S protein sequence in the full-length genome and DVG identified in this study may contribute to coronaviral fitness under diverse selection pressures.
-
-
-
Saliva sampling and its direct lysis is an excellent option for SARS-CoV-2 diagnosis in paediatric patients: comparison with the PanBio COVID-19 antigen rapid test in symptomatic and asymptomatic children
More LessIntroduction. Lateral flow test (LFTs) have been used as an alternative to reverse transcription quantitative PCR (RT-qPCR) in point-of-care testing. Despite their benefits, the sensitivity of LFTs may be low and is affected by several factors. We have previously reported the feasibility of using direct lysis of individual or pools of saliva samples from symptomatic and asymptomatic patients as a source of viral genomes for detection by RT-qPCR.
Hypothesis. Direct lysed saliva is more sensitive than antigen tests to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in samples from children.
Aim. Our goals here were to valuate the specificity and sensitivity of the PanBio COVID-19 antigen rapid test device (Ag-RTD) compared with RT-qPCR of direct lysed saliva.
Methodology. We evaluated the performance of the PanBio COVID-19 Ag-RTD in comparison to RT-qPCR direct lysed saliva from paired samples of 256 symptomatic and 242 asymptomatic paediatric patients.
Results. Overall, although there were no differences in the specificity (96.6%), we found a lower sensitivity (64.3%) of the PanBio Ag-test RTD compared to saliva in both symptomatic and asymptomatic patients. In addition, the sensitivity of PanBio was not correlated with the viral load present in the samples.
Conclusion. Our data highlight the benefits of using RT-qPCR and saliva samples for SARS-CoV-2 detection, particularly in paediatric patients.
-
-
-
Desmoglein-2 and COVID-19 complications: insights into its role as a biomarker, pathogenesis and clinical implications
More LessDesmoglein-2 (DSG2) has emerged as a potential biomarker for coronavirus disease 2019 (COVID-19) complications, particularly cardiac and cardiovascular involvement. The expression of DSG2 in lung tissues has been detected at elevated levels, and circulating DSG2 levels correlate with COVID-19 severity. DSG2 may contribute to myocardial injury, cardiac dysfunction and vascular endothelial dysfunction in COVID-19. Monitoring DSG2 levels could aid in risk stratification, early detection and prognostication of COVID-19 complications. However, further research is required to validate DSG2 as a biomarker. Such research will aim to elucidate its precise role in pathogenesis, establishing standardized assays for its measurement and possibly identifying therapeutic targets.
-
-
-
Novel neutralizing mouse-human chimeric monoclonal antibodies against the SARS-CoV-2 receptor binding domain
More LessIntroduction. Neutralizing antibodies have been widely used for the prophylaxis and treatment of COVID-19.
Hypothesis. The major target for these neutralizing antibodies is the receptor-binding domain (RBD) of the viral spike protein.
Aim. In the present study, we developed and characterized three neutralizing chimeric mouse-human mAbs for potential therapeutic purposes.
Methodology. Light and heavy chain variable region genes of three mouse mAbs (m4E8, m3B6, and m1D1) were amplified and ligated to human Cγ1 and Cκ constant region genes by PCR. After cloning into a dual promoter mammalian expression vector, the final constructs were transiently expressed in DG-44 cells and the purified chimeric antibodies were characterized by ELISA and Western blotting. The neutralizing potency of the chimeric mAbs was determined by three different virus neutralization tests including sVNT, pVNT, and cVNT.
Results. All three recombinant chimeric mAbs display human constant regions and are able to specifically bind to the RBD of SARS-CoV-2 with affinities comparable to the parental mAbs. Western blot analysis showed similar epitope specificity profiles for both the chimeric and the parental mouse mAbs. The results of virus neutralization tests (sVNT, pVNT, and cVNT) indicate that c4E8 had the most potent neutralizing activity with IC50 values of 1.772, 0.009, and 0.01 µg ml−1, respectively. All chimeric and mouse mAbs displayed a similar pattern of reactivity with the spike protein of the SARS-CoV-2 variants of concern (VOC) tested, including alpha, delta, and wild-type.
Conclusion. The chimeric mAbs displayed neutralizing potency similar to the parental mouse mAbs and are potentially valuable tools for disease control.
-
-
-
SARS-CoV-2 in outdoor air following the third wave lockdown release, Portugal, 2021
More LessAiming to contribute with more data on the presence of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) in outdoor environments, we performed air sampling in outdoor terraces from restaurants in three major cities of Portugal in April 2021, following the third wave lockdown release in the country. Air samples (n=19) were collected in 19 restaurant terraces during lunch time. Each air sample was collected using a Coriolis Compact air sampler, followed by RNA extraction and real-time quantitative PCR for the detection of viral RNA. Viral viability was also assessed through RNAse pre-treatment of samples. Only one of the 19 air samples was positive for SARS-CoV-2 RNA, with 7337 gene copies m–3 for the genomic region N2, with no viable virus in this sample. The low number of positive samples found in this study is not surprising, as sampling took place in outdoor settings where air circulation is optimal, and aerosols are rapidly dispersed by the air currents. These results are consistent with previous reports stating that transmission of SARS-CoV-2 in outdoor spaces is low, although current evidence shows an association of exposures in settings where drinking and eating is possible on-site with an increased risk in acquiring SARS-CoV-2 infection. Moreover, the minimal infectious dose for SARS-CoV-2 still needs to be determined so that the real risk of infection in different environments can be accurately established.
-
-
-
Performance evaluation of Novaplex SARS-CoV-2 variants assay kit series for SARS-CoV-2 detection using single nucleotide polymorphisms
More LessSevere acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants have received increasing attention globally because of their increased transmissibility and potential to escape immunity. Although whole-genome sequencing is the gold standard method for SARS-CoV-2 mutation detection and lineage determination, it is costly and time-consuming. However, SARS-CoV-2 variants can be identified based on select variant-specific single nucleotide polymorphisms (SNPs) in the spike protein-encoding gene (S). This study validated and compared the limit of detection (LOD) of L452R, N501Y, HV69/70 del and E484K as variant-specific SNPs of the S gene and RdRP as a SARS-CoV-2-specific gene, using the Novaplex SARS-CoV-2 variants assay kit series. For three SARS-CoV-2 lineages (B.1.617.2, B.1.1.7 and R.1), one strain per lineage was used. Variant-specific SNPs of the S gene were analysed using the Novaplex SARS-CoV-2 variants I assay and Novaplex SARS-CoV-2 variants II assay kits. Validation confirmed the LODs of the variant kits. The LOD for each target variant-specific SNP and RdRP was five RNA copies per reaction. The Novaplex SARS-CoV-2 variants assay kit series performs well and the LOD for SARS-CoV-2 detection and variant-specific SNP detection are consistent. The kits are suitable for use as routine laboratory tests for SARS-CoV-2 and variant-specific SNP detection in a single step, saving time and labour.
-
-
-
SARS-CoV-2 seroprevalence in hospital healthcare workers in Western Switzerland at the end of the second pandemic wave
More LessIntroduction. In early January 2020, the pandemic of COVID-19 (coronavirus disease 2019) rapidly spread from China and caused a worldwide pandemic.
Hypothesis. Healthcare workers represent a high-risk group for acquiring COVID-19 and for nosocomial transmission of severe acute respiratory coronavirus 2 (SARS-CoV-2).
Aim. We aimed to investigate over a 1 year period, across two pandemic waves, the SARS-CoV-2 seroprevalence in employees at a Western Switzerland public hospital.
Methodology. A prospective observational SARS-CoV-2 seroprevalence study was proposed to all hospital employees who enrolled on a voluntary basis.
Results. Out of 594 participants recruited on a voluntary basis, 269 volunteers (45.3 %) had anti-SARS-CoV-2 antibodies: this seroprevalence was twice higher than that reported in the local community. Healthcare workers with prolonged exposure to patients with COVID-19 showed a significantly higher odds ratio (OR) of having a positive SARS-CoV-2 serology [OR 3.19, 95 % confidence interval (CI) 2.16–4.74]. Symptoms showing the highest association with a positive serology were anosmia (OR 11.9, 95 % CI 5.58–30.9) and ageusia (OR 10.3, 95 % CI 4.8–26.3). A total of 17.1 % (95 % CI 12.2–21.1 %) of SARS-CoV-2 seropositive volunteers did not report a suspicion of COVID-19 in their personal history.
Conclusion. Overall, we observed that the impact of the second SARS-CoV-2 pandemic wave was considerable and significantly affected healthcare workers with prolonged exposure to patients with COVID-19.
-
-
-
The acceptability of testing contacts of confirmed COVID-19 cases using serial, self-administered lateral flow devices as an alternative to self-isolation
More LessIntroduction. Evidence suggests that although people modify their behaviours, full adherence to self-isolation guidance in England may be suboptimal, which may have a detrimental impact on COVID-19 transmission rates.
Hypothesis. Testing asymptomatic contacts of confirmed COVID-19 cases for the presence of SARS-CoV-2 could reduce onward transmission by improving case ascertainment and lessen the impact of self-isolation on un-infected individuals.
Aim. This study investigated the feasibility and acceptability of implementing a ‘test to enable approach’ as part of England’s tracing strategy.
Methodology. Contacts of confirmed COVID-19 cases were offered serial testing as an alternative to self-isolation using daily self-performed lateral flow device (LFD) tests for the first 7 days post-exposure. Asymptomatic participants with a negative LFD result were given 24 h of freedom from self-isolation between each test. A self-collected confirmatory PCR test was performed on testing positive or at the end of the LFD testing period.
Results. Of 1760 contacts, 882 consented to daily testing, of whom 812 individuals were within 48 h of exposure and were sent LFD testing packs. Of those who declined to participate, 39.1% stated they had already accessed PCR testing. Of the 812 who were sent LFD packs, 570 (70.2%) reported one or more LFD results; 102 (17.9%) tested positive. Concordance between reported LFD result and a supplied LFD image was 97.1%. In total, 82.8% of PCR-positive samples and 99.6% of PCR-negative samples were correctly detected by LFD. The proportion of secondary cases from contacts of those who participated in the study and tested positive (6.3%; 95% CI: 3.4–11.1%) was comparable to a comparator group who self-isolated (7.6%; 95% CI: 7.3–7.8%).
Conclusion. This study shows a high acceptability, compliance and positivity rates when using self-administered LFDs among contacts of confirmed COVID-19 cases. Offering routine testing as a structured part of the contact tracing process is likely to be an effective method of case ascertainment.
-
-
-
Replacement of the Alpha variant of SARS-CoV-2 by the Delta variant in Lebanon between April and June 2021
More LessGeorgi Merhi, Alexander J. Trotter, Leonardo de Oliveira Martins, Jad Koweyes, Thanh Le-Viet, Hala Abou Naja, Mona Al Buaini, Sophie J. Prosolek, Nabil-Fareed Alikhan, Martin Lott, Tatiana Tohmeh, Bassam Badran, Orla J. Jupp, Sarah Gardner, Matthew W. Felgate, Kate A. Makin, Janine M. Wilkinson, Rachael Stanley, Abdul K. Sesay, Mark A. Webber, Rose K. Davidson, Nada Ghosn, Mark Pallen, Hamad Hasan, Andrew J. Page and Sima TokajianThe COVID-19 pandemic continues to expand globally, with case numbers rising in many areas of the world, including the Eastern Mediterranean Region. Lebanon experienced its largest wave of COVID-19 infections from January to April 2021. Limited genomic surveillance was undertaken, with just 26 SARS-CoV-2 genomes available for this period, nine of which were from travellers from Lebanon detected by other countries. Additional genome sequencing is thus needed to allow surveillance of variants in circulation. In total, 905 SARS-CoV-2 genomes were sequenced using the ARTIC protocol. The genomes were derived from SARS-CoV-2-positive samples, selected retrospectively from the sentinel COVID-19 surveillance network, to capture diversity of location, sampling time, sex, nationality and age. Although 16 PANGO lineages were circulating in Lebanon in January 2021, by February there were just four, with the Alpha variant accounting for 97 % of samples. In the following 2 months, all samples contained the Alpha variant. However, this had changed dramatically by June and July 2021, when all samples belonged to the Delta variant. This study documents a ten-fold increase in the number of SARS-CoV-2 genomes available from Lebanon. The Alpha variant, first detected in the UK, rapidly swept through Lebanon, causing the country's largest wave to date, which peaked in January 2021. The Alpha variant was introduced to Lebanon multiple times despite travel restrictions, but the source of these introductions remains uncertain. The Delta variant was detected in Gambia in travellers from Lebanon in mid-May, suggesting community transmission in Lebanon several weeks before this variant was detected in the country. Prospective sequencing in June/July 2021 showed that the Delta variant had completely replaced the Alpha variant in under 6 weeks.
-
-
-
Catwalk: identifying closely related sequences in large microbial sequence databases
More LessThere is a need to identify microbial sequences that may form part of transmission chains, or that may represent importations across national boundaries, amidst large numbers of SARS-CoV-2 and other bacterial or viral sequences. Reference-based compression is a sequence analysis technique that allows both a compact storage of sequence data and comparisons between sequences. Published implementations of the approach are being challenged by the large sample collections now being generated. Our aim was to develop a fast software detecting highly similar sequences in large collections of microbial genomes, including millions of SARS-CoV-2 genomes. To do so, we developed Catwalk, a tool that bypasses bottlenecks in the generation, comparison and in-memory storage of microbial genomes generated by reference mapping. It is a compiled solution, coded in Nim to increase performance. It can be accessed via command line, rest api or web server interfaces. We tested Catwalk using both SARS-CoV-2 and Mycobacterium tuberculosis genomes generated by prospective public-health sequencing programmes. Pairwise sequence comparisons, using clinically relevant similarity cut-offs, took about 0.39 and 0.66 μs, respectively; in 1 s, between 1 and 2 million sequences can be searched. Catwalk operates about 1700 times faster than, and uses about 8 % of the RAM of, a Python reference-based compression and comparison tool in current use for outbreak detection. Catwalk can rapidly identify close relatives of a SARS-CoV-2 or M. tuberculosis genome amidst millions of samples.
-
-
-
Identification and characterization of virus-encoded circular RNAs in host cells
More LessEmerging evidence has identified viral circular RNAs (circRNAs) in human cells infected by viruses, interfering with the immune system and inducing diseases including human cancer. However, the biogenesis and regulatory mechanisms of virus-encoded circRNAs in host cells remain unknown. In this study, we used the circRNA detection tool CIRI2 to systematically determine the virus-encoded circRNAs in virus-infected cancer cell lines and cancer patients, by analysing RNA-Seq datasets derived from RNase R-treated samples. Based on the thousands of viral circRNAs we identified, the biological characteristics and potential roles of viral circRNAs in regulating host cell function were determined. In addition, we developed a Viral-circRNA Database (http://www.hywanglab.cn/vcRNAdb/), which is open to all users to search, browse and download information on circRNAs encoded by viruses upon infection.
-
-
-
Evaluation of the antifungal effect of chlorogenic acid against strains of Candida spp. resistant to fluconazole: apoptosis induction and in silico analysis of the possible mechanisms of action
More LessCecília Rocha da Silva, Lívia Gurgel do Amaral Valente Sá, Ermerson Vieira dos Santos, Thais Lima Ferreira, Tatiana do Nascimento Paiva Coutinho, Lara Elloyse Almeida Moreira, Rosana de Sousa Campos, Claudia Roberta de Andrade, Wildson Max Barbosa da Silva, Igor de Sá Carneiro, Jacilene Silva, Hélcio Silva dos Santos, Emmanuel Silva Marinho, Bruno Coelho Cavalcanti, Manoel Odorico de Moraes, Hélio Vitoriano Nobre Júnior and João Batista Andrade NetoIntroduction. Candida spp. are commensal fungal pathogens of humans, but when there is an imbalance in the microbiota, or weak host immunity, these yeasts can become pathogenic, generating high medical costs.
Gap Statement. With the increase in resistance to conventional antifungals, the development of new therapeutic strategies is necessary.
This study evaluated the in vitro antifungal activity of chlorogenic acid against fluconazole-resistant strains of Candida spp.
Mechanism of action through flow cytometry and in silico analyses, as well as molecular docking assays with ALS3 and SAP5, important proteins in the pathogenesis of Candida albicans associated with the adhesion process and biofilm formation.
Results. The chlorogenic acid showed in vitro antifungal activity against the strains tested, causing reduced cell viability, increased potential for mitochondrial depolarization and production of reactive oxygen species, DNA fragmentation and phosphatidylserine externalization, indicating an apoptotic process. Concerning the analysis through docking, the complexes formed between chlorogenic acid and the targets Thymidylate Kinase, CYP51, 1Yeast Cytochrome BC1 Complex e Exo-B-(1,3)-glucanase demonstrated more favourable binding energy. In addition, chlorogenic acid presented significant interactions with the ALS3 active site residues of C. albicans, important in the adhesion process and resistance to fluconazole. Regarding molecular docking with SAP5, no significant interactions were found between chlorogenic acid and the active site of the enzyme.
Conclusion. We concluded that chlorogenic acid has potential use as an adjuvant in antifungal therapies, due to its anti-Candida activity and ability to interact with important drug targets.
-
-
-
Network pharmacology and experimental validation identify the potential mechanism of sophocarpine for COVID-19
More LessIntroduction. Coronavirus disease 2019 (COVID-19) has caused a serious threat to public health worldwide, and there is currently no effective therapeutic strategy for treating COVID-19.
Hypothesis/Gap Statement. We propose that sophocarpine (SOP) might have potential therapeutic effects on COVID-19 through inhibiting the cytokine storm and the nuclear factor NF-κB signalling pathway.
Aim. The objective was to elucidate the potential mechanism of SOP against COVID-19 through a network pharmacology analysis and its experimental validation.
Methodology. The BATMAN-TCM database was used to identify the therapeutic targets of SOP, while the GeneCards and DisGeNET databases were used to identify the targets related to COVID-19. A protein–protein interaction (PPI) network was constructed from the STRING and analysed using Cytoscape software. Gene ontology (GO), Kyoto Encyclopaedia of Genes and Genomes (KEGG) and disease ontology (DO) enrichment analyses of the co-targets were performed using Metascape. Autodock 4.2.6 and Pymol software were applied for molecular docking. Levels of the proinflammatory cytokines IL-6, TNFα and IL-1β were measured by ELISA, while mRNA expression levels of intercellular adhesion molecule 1 (ICAM-1), vascular endothelial growth factor A (VEGFA) and IFN gamma (IFNG) were detected by real-time quantitative reverse transcription PCR. The protein levels of the molecules involved in the NF-κB signalling pathway were validated by western blot analysis.
Results. A total of 65 co-targets of SOP and COVID-19 were determined. GO and KEGG enrichment analyses suggested that SOP affected COVID-19 by regulating the IL-17 signalling pathway, TNF signalling pathway and other signalling pathways. The PPI network and molecular docking showed that p65, ICAM-1 and VEGFA were key targets of SOP against COVID-19 and the underlying mechanism was validated in A549 cells in vitro. SOP attenuated the LPS-induced production of TNF-α and IL-6 and downregulated the LPS-induced mRNA expression of ICAM-1, VEGFA and IFNG. Mechanistically, SOP pretreatment inhibited the phosphorylation of p65 and facilitated the activation of Nrf2.
Conclusions. SOP has a potential therapeutic effect on COVID-19 through multiple pathways and targets, and inhibits the production of pro-inflammatory cytokines and molecules involved in the NF-κB signalling pathway.
-
-
-
Reporting of RT-PCR cycle threshold (Ct) values during the first wave of COVID-19 in Qatar improved result interpretation in clinical and public health settings
More LessPeter V. Coyle, Naema Hassan Al Molawi, Mohamed Ali Ben Hadj Kacem, Reham Awni El Kahlout, Einas Al Kuwari, Abdullatif Al Khal, Imtiaz Gillani, Andrew Jeremijenko, Hatoun Saeb, Mohammad Al Thani, Roberto Bertollini, Hanan F. Abdul Rahim, Hiam Chemaitelly, Patrick Tang, Ali Nizar Latif, Saad Al Kaabi, Muna A. Rahman S. Al Maslamani, Brendan David Morris, Nasser Al-Ansari, Anvar Hassan Kaleeckal and Laith J. Abu RaddadIntroduction. The cycle threshold (Ct) value in real-time PCR (RT-PCR) is where a target-specific amplification signal becomes detectable and can infer viral load, risk of transmission and recovery. Use of Ct values in routine practice is uncommon.
Gap Statement. There is a lack of routine use of Ct values when reporting RT-PCR results in routine practice.
Aim. To automatically insert Ct values and interpretive comments when reporting SARS-CoV-2 RT-PCR to improve patient management.
Methodology. Routine Ct values across three different RT-PCR platforms were reviewed for concordance at presentation and clearance in patients with COVID-19. An indicative threshold (IT) linked to viral clearance kinetics was defined at Ct30 to categorize Ct values as low and high, reflecting high and low viral loads respectively.
Results. The different gene targets of each platform showed high correlation and kappa score agreement (P<0.001). Average Ct values were automatically generated with values ≤Ct30 reported as positive and >Ct30 as reactive; interpretive comments were added to all reports. The new reporting algorithm impacted on: physician interpretation of SARS-CoV-2 results; patient management and transfer; staff surveillance; length of stay in quarantine; and redefinition of patient recovery.
Conclusion. Incorporation of Ct values into routine practice is possible across different RT-PCR platforms and adds useful information for patient management. The use of an IT with interpretive comments improves clinical interpretation and could be a model for reporting other respiratory infections. Withholding Ct values wastes useful clinical data and should be reviewed by the profession, accreditation bodies and regulators.
-
-
-
SARS-CoV-2 and Prevotella spp.: friend or foe? A systematic literature review
More LessDuring this global pandemic of the COVID-19 disease, a lot of information has arisen in the media and online without scientific validation, and among these is the possibility that this disease could be aggravated by a secondary bacterial infection such as Prevotella, as well as the interest or not in using azithromycin, a potentially active antimicrobial agent. The aim of this study was to carry out a systematic literature review, to prove or disprove these allegations by scientific arguments. The search included Medline, PubMed, and Pubtator Central databases for English-language articles published 1999–2021. After removing duplicates, a total of final eligible studies (n=149) were selected. There were more articles showing an increase of Prevotella abundance in the presence of viral infection like that related to Human Immunodeficiency Virus (HIV), Papillomavirus (HPV), Herpesviridae and respiratory virus, highlighting differences according to methodologies and patient groups. The arguments for or against the use of azithromycin are stated in light of the results of the literature, showing the role of intercurrent factors, such as age, drug consumption, the presence of cancer or periodontal diseases. However, clinical trials are lacking to prove the direct link between the presence of Prevotella spp. and a worsening of COVID-19, mainly those using azithromycin alone in this indication.
-
-
-
A comparison of SARS-CoV-2 RNA extraction with the QuickGene-810 Nucleic Acid Isolation System compared to the EZ1 Advanced DSP Virus Kit
More LessThe QuickGene-810 Nucleic Acid Isolation System is a semi-automated extraction platform which may be used for RNA extraction. New methods were required to support the rapid increase in respiratory virus testing during the SARS-CoV-2 pandemic. The aim of this study was to assess SARS-CoV-2 RNA extraction using the QuickGene-810 kit compared to the EZ1 Advanced Extraction Platform for use on the AusDiagnostics SARS-CoV-2, Influenza and RSV 8-well RT-PCR assay. Qualitative results from all clinical samples were concordant between the QuickGene-810 and the EZ1 extraction methods, demonstrating that the QuickGene-810 kit is suitable for use in pathogen diagnostics. However, there was an average difference of approximately two cycles between the cycle threshold (Ct) values for both SARS-CoV-2 targets, suggesting that the EZ1 kit yields a higher concentration of nucleic acid extract, possibly related to its use of carrier RNA and/or smaller elution volume, which infers the possibility of false negative results for samples with very low viral loads.
-
-
-
Feasibility of a refurbished shipping container as a transportable laboratory for rapid SARS-CoV-2 diagnostics
More LessBackground. Australia’s response to the coronavirus disease 2019 (COVID-19) pandemic relies on widespread availability of rapid, accurate testing and reporting of results to facilitate contact tracing. The extensive geographical area of Australia presents a logistical challenge, with many of the population located distant from a laboratory capable of robust severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. A strategy to address this is the deployment of a mobile facility utilizing novel diagnostic platforms. This study aimed to evaluate the feasibility of a fully contained transportable SARS-CoV-2 testing laboratory using a range of rapid point-of-care tests.
Method. A 20 ft (6.1 m) shipping container was refurbished (GeneWorks, Adelaide, South Australia) with climate controls, laboratory benches, hand-wash station and a class II biosafety cabinet. Portable marquees situated adjacent to the container served as stations for registration, sample acquisition and personal protective equipment for staff. Specimens were collected and tested on-site utilizing either the Abbott ID NOW or Abbott Panbio rapid tests. SARS-CoV-2 positive results from the rapid platforms or any participants reporting symptoms consistent with COVID-19 were tested on-site by GeneXpert Xpress RT-PCR. All samples were tested in parallel with a standard-of-care RT-PCR test (Panther Fusion SARS-CoV-2 assay) performed at the public health reference laboratory. In-laboratory environmental conditions and data management-related factors were also recorded.
Results. Over a 3 week period, 415 participants were recruited for point-of-care SARS-CoV-2 testing. From time of enrolment, the median result turnaround time was 26 min for the Abbott ID NOW, 32 min for the Abbott Panbio and 75 min for the Xpert Xpress. The environmental conditions of the refurbished shipping container were found to be suitable for all platforms tested, although humidity may have produced condensation within the container. Available software enabled turnaround times to be recorded, although technical malfunction resulted in incomplete data capture.
Conclusion. Transportable container laboratories can enable rapid COVID-19 results at the point of care and may be useful during outbreak settings, particularly in environments that are physically distant from centralized laboratories. They may also be appropriate in resource-limited settings. The results of this pilot study confirm feasibility, although larger trials to validate individual rapid point-of-care testing platforms in this environment are required.
-
-
-
SARS-CoV-2 variants of concern alpha, beta, gamma and delta have extended ACE2 receptor host ranges
More LessFollowing the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in PR China in late 2019 a number of variants have emerged, with two of these – alpha and delta – subsequently growing to global prevalence. One characteristic of these variants are changes within the spike protein, in particular the receptor-binding domain (RBD). From a public health perspective, these changes have important implications for increased transmissibility and immune escape; however, their presence could also modify the intrinsic host range of the virus. Using viral pseudotyping, we examined whether the variants of concern (VOCs) alpha, beta, gamma and delta have differing host angiotensin-converting enzyme 2 (ACE2) receptor usage patterns, focusing on a range of relevant mammalian ACE2 proteins. All four VOCs were able to overcome a previous restriction for mouse ACE2, with demonstrable differences also seen for individual VOCs with rat, ferret or civet ACE2 receptors, changes that we subsequently attributed to N501Y and E484K substitutions within the spike RBD.
-
-
-
Second infection with SARS-CoV-2 wild-type is associated with increased disease burden after primary SARS-CoV-2/HBoV-1 coinfection, Cologne, Germany
More LessSARS-CoV-2 is the cause of the still-ongoing COVID-19 pandemic. To date reports on re-infections after full recovery from a previous COVID-19 course remain limited due to the fact that re-infections or second infections occur at the earliest between 3 to 24 months after full recovery while the pandemic lasts only since a year. Even less data are available on re-infections associated with emerging variants.
A 33-year-old previously healthy male patient was tested twice SARS-CoV-2 RNA positive with an 8 months symptom-free interval between the two COVID-19 episodes in our setting in Cologne, Germany. While the first episode was accompanied by a co-detection of human bocavirus and hardly any symptoms, the second episode was characterized by serious illness and severe flu-like symptoms, although hospitalization was not required. After the first episode no residual viral RNA was detected after the patient was released from quarantine. Follow up of the patient revealed a moderate but significant reduction of the lung volume and slightly impaired diffusion capacity.
Conclusion. While it is known that re-infections with SARS-CoV-2 may occur this is the first report of a co-detection of human bocavirus (HBoV) during a primary SARS-CoV-2 infection. The first, hardly symptomatic episode showed that co-infections do not necessarily initiate severe COVID-19 courses. The second more severe episode with serious flu-like symptoms could be explained by the sustained mild damage of the airways during the primary infection.
-
-
-
An isothermal amplification-coupled dipstick for the rapid detection of COVID-19
More LessEarly detection of coronavirus disease 2019 (COVID-19) is critical for both initiating appropriate treatment and preventing the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent. A simple and rapid diagnostic test that can be performed without any expensive equipment would be valuable for clinicians working in a low-resource setting. Here, we report a point-of-care detection technique for COVID-19 that combines the power of isothermal amplification (reverse transcription helicase-dependent amplification, RT-HDA) and dipstick technologies. The limit of detection of this diagnostic test is six copies of SARS-CoV-2 µl−1 in clinical specimens. Of the 22 clinical specimens tested, RT-HDA-coupled dipstick correctly identified all positive and negative specimens. The RT-HDA can be performed over a heating block and the results can be interpreted visually with the dipstick technology without any specialized equipment. Furthermore, the RT-HDA-coupled dipstick could be performed in a short turnaround time of ~2 h. Thus, the RT-HDA-coupled dipstick could serve as a point-of-care diagnostic test for COVID-19 in a low-resource environment.
-
