Collections
Browse our Collections – bringing together peer-reviewed content from across the Society’s publishing platform on a range of hot topics and subject areas.
1 - 50 of 56 results
-
-
Anaerobe
More Less
Anaerobic clinical microbiology remains a challenge due to specialist culture requirements, coupled with the increase in and spread of antimicrobial resistance. The normal human microbiota is primarily composed of anaerobic bacteria, and is now recognised as a source of life-threatening anaerobic infection. More recent metataxonomic and metagenomic sequencing has extended interest in the potential role of the microbiota in a plethora of other aspects of human health, from obesity to mental health. In addition, the successful use of faecal microbiota transplants for the treatment of clostridial infection raises potential unchartered long-term consequences and possibilities.
In conjunction with Anaerobe 2019 and Anaerobe 2021, this collection will provide scientific insights into the future impact of anaerobic bacteria in human health and disease, addressing the implications of recent microbiota studies as well as the continued threat of emerging and re-emerging anaerobic infection.
This collection is open for submissions – please submit your article here, stating that your manuscript is part of the Anaerobe collection.
-
-
-
Antimicrobial Efflux
More Less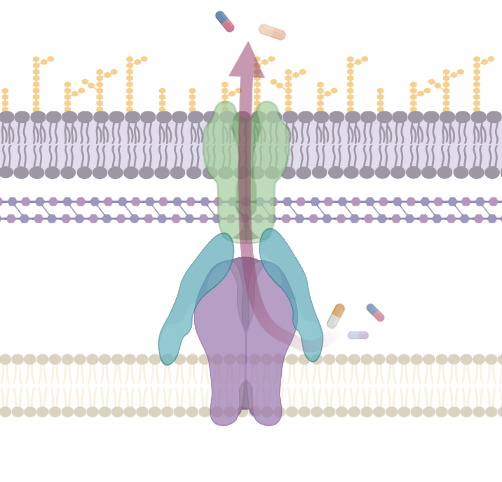
Bacterial efflux pumps are a complex and diverse set of membrane proteins responsible for transport of substrates between cellular compartments or to the outside of bacterial cells. They are important mediators of antimicrobial resistance and are critical for many bacterial pathogens to cause infection and form biofilm. This collection has been put together to mark two very special anniversaries. It is 40 years since George and Levy (1983) published the discovery of the first 'energy-dependent efflux system' conferring resistance to Tetracycline and 30 years since the first description of the pumps from the RND family (Poole et al., 1993 and Ma et al., 1993). In this time our understanding of efflux function, physiology and structure have been transformed. This special collection is guest edited by Drs Jessica Blair and Ayush Kumar and aims to highlight the diversity of efflux pump research and the most recent advances in this fascinating field. We also invite articles describing advances in methodology that aid the characterization of efflux pumps. This collection is open for submissions in Microbiology. Upon submission, please indicate that your manuscript is to be considered for the Antimicrobial Efflux collection.
-
-
-
Antimicrobial Resistance
More Less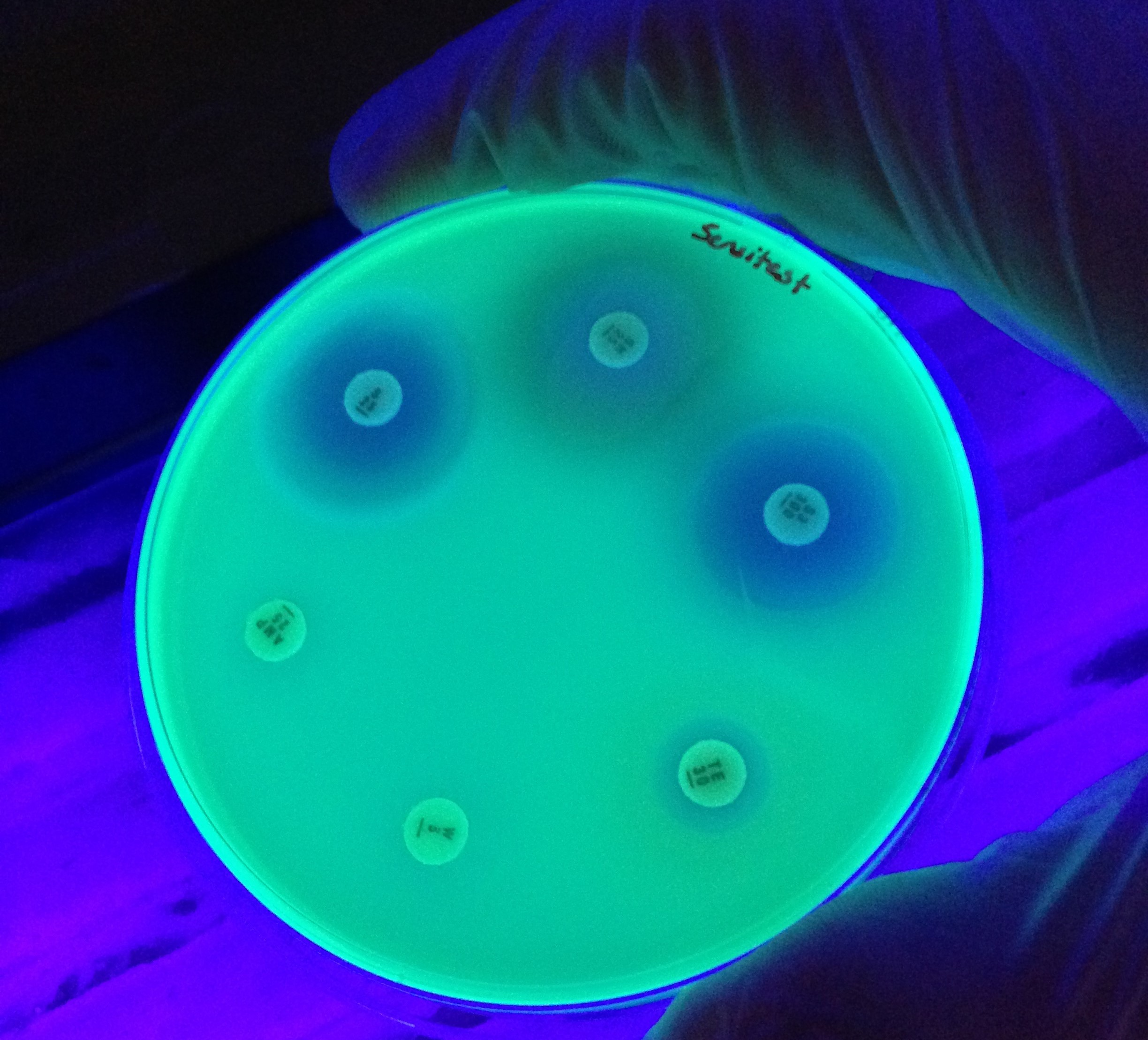
Antimicrobial drugs underpin much of modern medicine. Bacteria, fungi, parasites and viruses that exhibit resistance to antimicrobials threaten the efficacy of therapeutics and impose significant global healthcare and economic burdens. In the midst of the ongoing COVID-19 pandemic, the emergence of pan-drug resistant bacterial and fungal infections in hospitals is a stark reminder of the looming antimicrobial resistance crisis.
That bacteria would develop resistance to antibiotics through mutation was predicted as early as the discovery of penicillin, but the observation of resistance gene transfer between strains in the 1950s heralded the beginning of a period of rapid discovery of resistance mechanisms. Research into the mechanisms of antibiotic resistance proceeded alongside the development of the field of molecular biology and the decades since have seen tremendous advances in our understanding. Still, there is much to learn about where, when and how multi-drug resistance evolves, which organisms are involved and how chains of transmission might be broken before clinicians are faced with intractable infections. Today, multidisciplinary research seeks to answer these questions and the latest advances in sequencing technologies and analysis capacities promise insights into the dynamics of resistance from single cells to complex microbial communities.
Guest-edited by Prof. Willem van Schaik and Dr. Robert Moran, this Antimicrobial Resistance special collection aims to highlight research on the emergence, accumulation and spread of antimicrobial resistance, with a particular focus on opportunistic pathogens and the mobile genetic elements therein.
-
-
-
Arboviruses and their Vectors
More Less
Curated by Journal of General Virology Editor Dr Eng Eong Ooi (Duke NUS Medical School) and Advisory Board Member Dr Esther Schnettler (Bernhard Nocht Institute for Tropical Medicine), this collection presents the latest advances in arbovirus research. This collection was launched in conjuction with IMAV 2017 and in line with IMAV 2019 welcomes submissions of original research articles, Insight Reviews and full-length Reviews.
Find out more about how to submit to the collection here.
-
-
-
Avian Infectious Diseases
More Less
Infectious diseases continue to threaten the sustainability, productivity and growth of the poultry industry worldwide and some present a risk to public health. Many are also present in wild bird populations, with the potential to spill over into domestic birds.
Previously curated by Professor Paul Britton and Dr Mike Skinner, in line with the ‘Pathogenesis and Molecular Biology of Avian Viruses’ Focused Meeting, this collection and the Focused Meeting have been expanded to include bacteria and parasites.
Guest-edited by organisers of the Avian Infectious Diseases 2021 Focused Meeting, Dr Holly Shelton (The Pirbright Institute) and Dr Andrew Broadbent (The Pirbright Institute and University of Maryland), this collection presents high-quality work from Journal of General Virology and Journal of Medical Microbiology on important avian pathogens: their interactions with the host, cell biology, molecular epidemiology and methods of control.This collection is open for submissions in both participating journals, Journal of General Virology and Journal of Medical Microbiology.
Image credit: iStock/Sonja Filitz.
-
-
-
Bacterial Cell Envelopes
More Less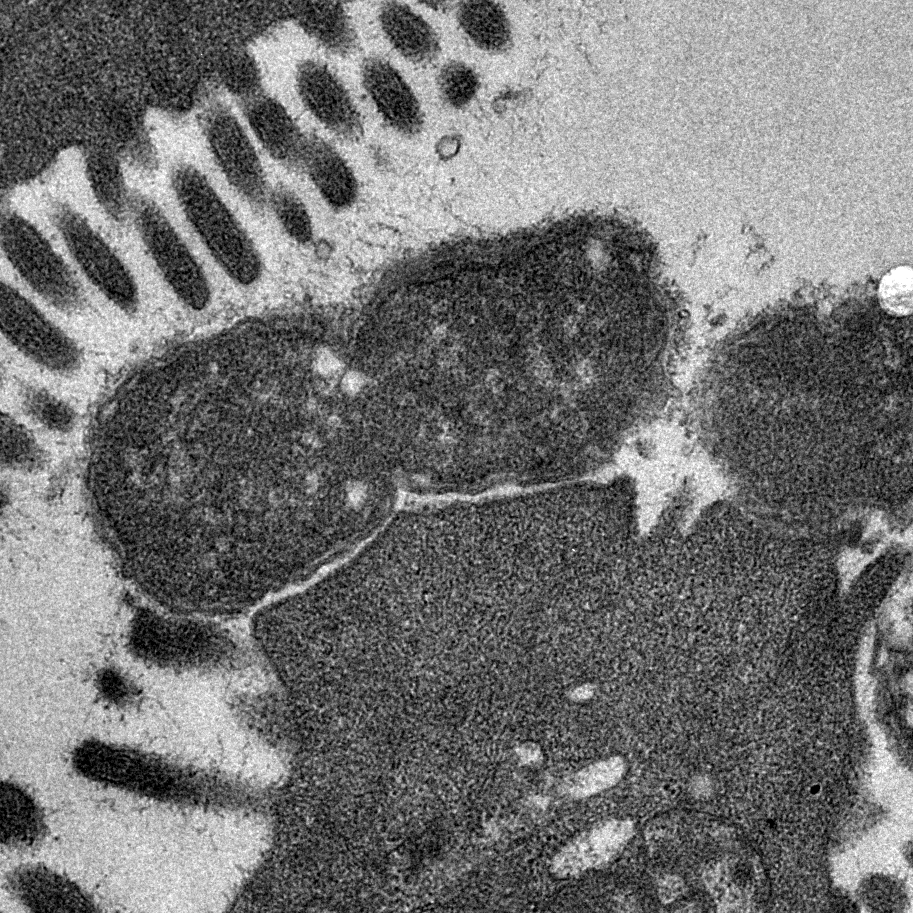 The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
The surfaces of bacteria are critically important. They form the first line of defence against external biotic and abiotic threats and are modified in many different ways to resist phagocytosis, phage predation and antimicrobial attack. Bacterial cell envelopes are selectively permeable to allow the uptake of important nutrients and to facilitate the export of waste products. Complex protein machines span across these structures to transport molecules that build and maintain the envelope. Bacteria often encode multiple different protein secretion systems that assemble surface structures such as pili and flagella. Many of these systems secrete proteins that mediate interactions with other living organisms. The essential nature of bacterial cell envelopes is reflected by the fact that they serve as targets for many of our most effective antibiotics.
Over the years, Microbiology has published many important findings that have contributed enormously to our understanding of the structure, function and biogenesis of bacterial envelopes. Here we celebrate the journal’s 75th year with a special collection of reviews guest-edited by Professor Tracy Palmer and Dr Yinka Somorin that highlights some of the most important areas of current research in this vibrant research field.
Image shows transmission electron micrograph of a dividing Citrobacter rodentium cell attached to infected mouse colonic epithelium. Characteristic ‘Attaching and Effacing lesions’ or raised pedestals can be seen beneath adhered C. rodentium, formed by the accumulation of host cell actin mediated by the bacterial type III protein secretion system. Effacement of the local brush border villi around the site of attachment is also observed. Courtesy of Dr James Connolly (Newcastle University).
-
-
-
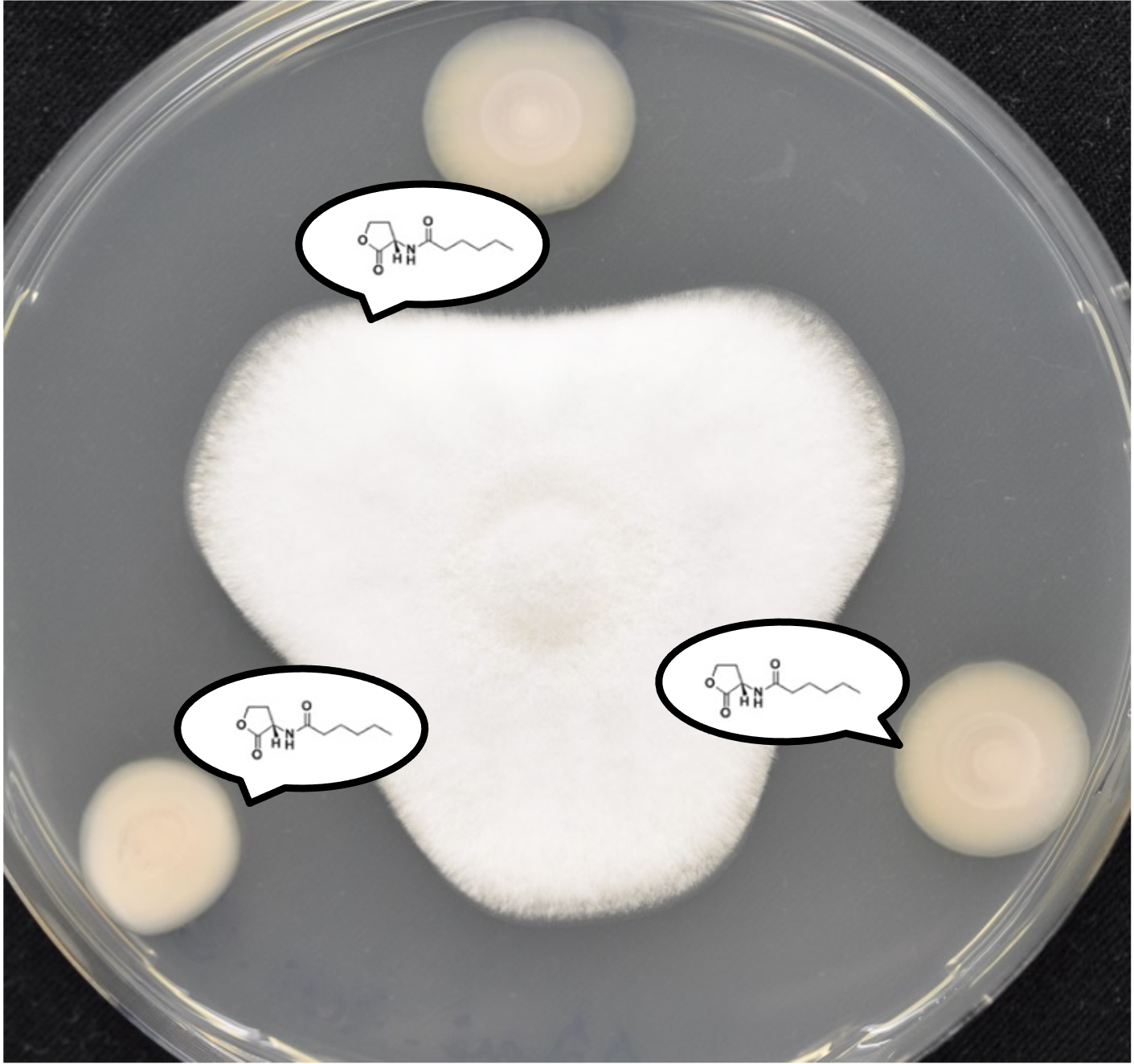
Bacterial Competitive Mechanisms
More Less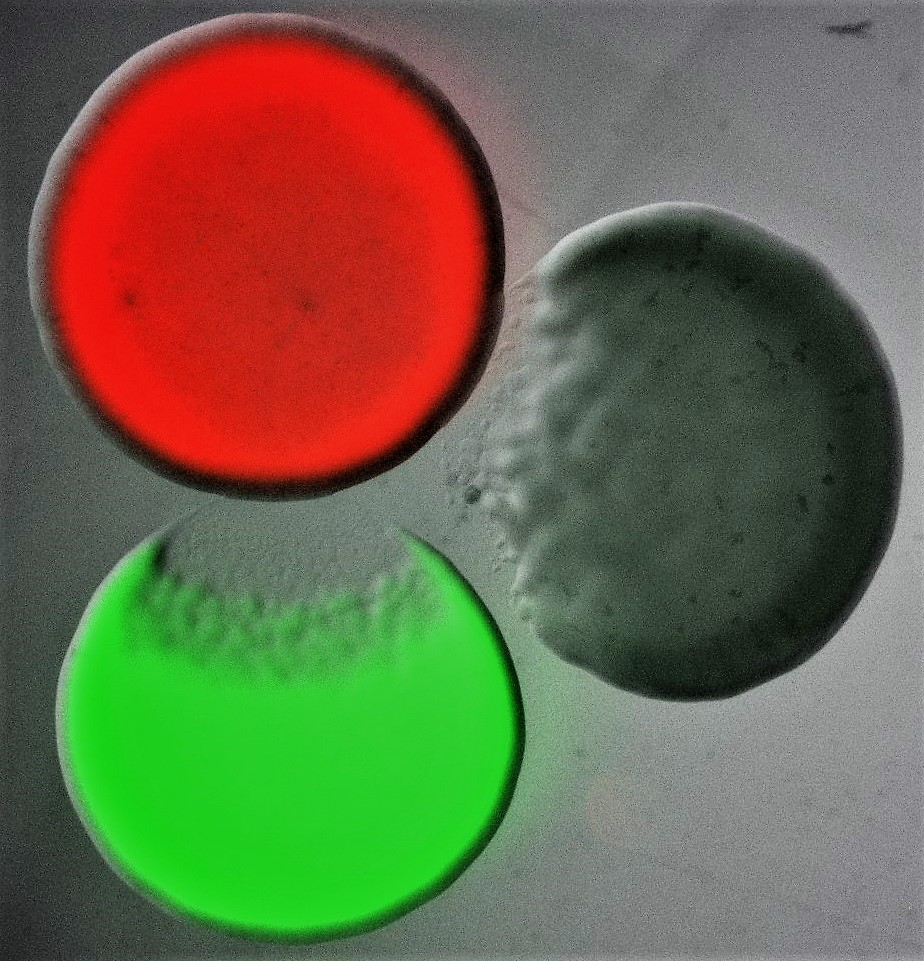
Bacteria are found in every environment and live in complex communities, which are characterized by large densities and diverse compositions. In these microbiomes, bacteria constantly compete for resources, such as nutrients, space and oxygen. Bacteria use a vast arsenal to eliminate their competitors, from elaborate contact-dependent nano-harpoons to small diffusible toxic compounds. In addition to producing these, oftentimes, elaborate weapons, bacteria also employ a range of collective behaviors that, in turn, allow them to carefully regulate weapon production, coordinate their attack strategies with their clonemates, and distinguish friends from foe. In essence, these microscopic warriors take advantage of complex regulation and interbacterial interaction mechanisms to balance the costs of weapon production and deployment with the benefits of gaining precious resources. As such, they manage to not only survive in their highly competitive microcosms, but to also build stable communities that are often crucial for the health of their animal, plant and human hosts.
Over the years, Microbiology has published many studies that have expanded our understanding of the competitive mechanisms bacteria use to thrive in their niches, including, but not limited to, the molecular characterization of toxins or secretion systems and the investigation of interbacterial competition at the organismal level. Here, we take the opportunity to celebrate the journal’s transition to fully Open Access with a special collection of articles that highlights some of the most important areas of current research in this vibrant research field. This collection is guest edited by Professor Despoina A.I. Mavridou (The University of Texas at Austin, USA), Professor Tracy Palmer (Newcastle University, UK) and Dr Sabrina L. Slater (The University of Texas at Austin, USA).
Image shows three differentially labelled Escherichia coli colonies, each of which is producing a different colicin toxin. Colicins are part of the bacteriocin toxin family, an ensemble of diffusible proteinaceous toxins that bacteria release, usually by lysis, to kill phylogenetically similar strains and species. Bacterial killing can be seen characteristically as an area of clearance at the edge of two of the three colonies. Courtesy of Despoina A.I. Mavridou (The University of Texas at Austin).
-
-
-
Bacteriophage
More LessBacteriophages were discovered 100 years ago and have since been a key tool used in biological research. Initially used as model organisms for work in genetics and molecular biology, bacteriophages are now known to be one of the major drivers of bacterial evolution and diversification. The introduction of sequencing technologies, phage genomics and metagenomics has highlighted their tremendous diversity and roles in controlling ecological systems within a range of environments. Due to their specificity, phage genomes are now also being manipulated as therapeutics, potentially providing an alternative to conventional antibiotics. Giant phages have also discovered and may represent a novel genus of living organism.Guest-edited by Professor Tetsuya Hayashi (Kyushu University), this collection brings together original Research Articles, Methods, Mini Reviews, and full-length Reviews relating to the diversity of bacteriophages and genomics-based research with a focus on their roles in the evolution of bacteria and ecosystems. Microbial Genomics also welcomes large-scale genomic analyses of bacteriophages which provide the basis for developing novel phage therapies.
This collection is now open for submissions. Submit here and state that your manuscript is part of the Bacteriophage Collection.
-
-
-
Candida
More Less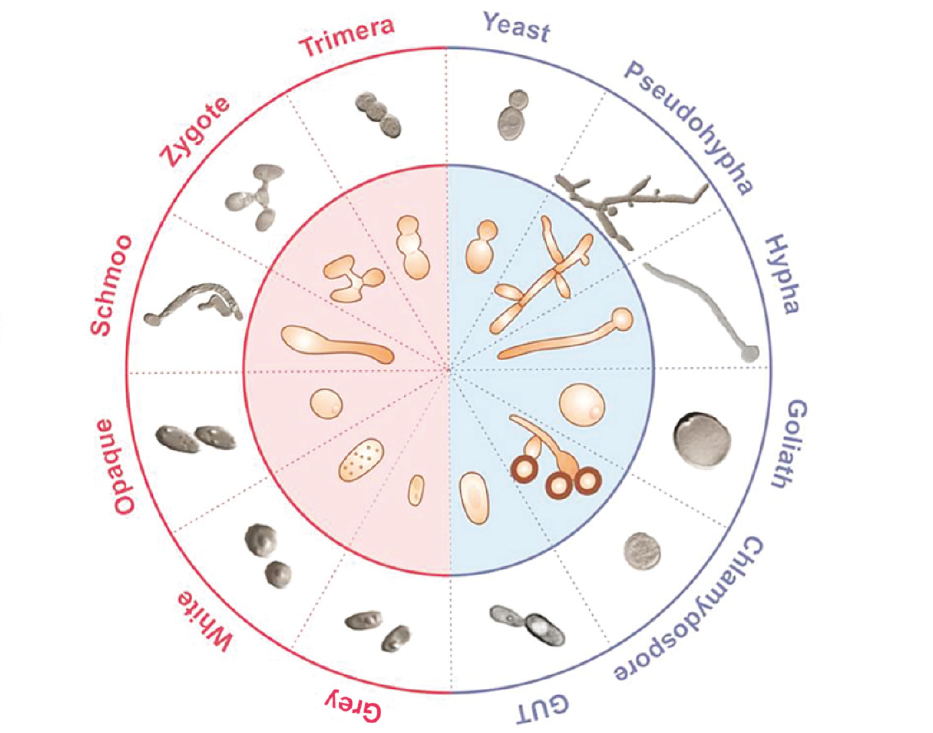
In conjunction with the Candida and Candidiasis meeting, Microbiology Society is bringing together research on Candida biology – species that are major causes of infectious disease in AIDS patients, cancer chemotherapy patients, premature infants, etc. The polymorphic yeast Candida albicans is the most important fungal pathogen in humans. Beyond the clinics, basic research in this organism deals with a variety of topics of interest, like the genetics and molecular biology behind antifungal drug resistance; the molecular determinants of endurance to nutritional, pH and oxidative stress imposed by the host defences; or its interactions with both the host epithelia and bacterial partners that share the mucosal microbiota with the fungus. This collection of research articles published on diverse aspects of Candida biology showcases the journals’ range of Candida research. This collection is open for submissions across our portfolio. Authors are invited to submit on any aspect of Candida research. Upon submission, please indicate that your manuscript is to be considered for the Candida collection.
-
-
-
Coronaviruses
More Less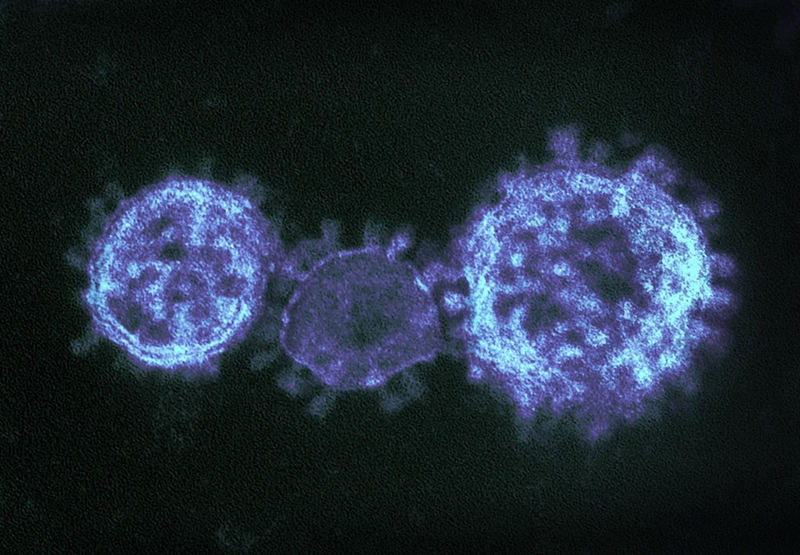
Coronaviruses are a large family of viruses that can infect a range of hosts. They are known to cause diseases including the common cold, Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) in humans.
In January 2020, China saw an outbreak of a new coronavirus strain now named SARS-CoV-2. Although the animal reservoir for the SARS and MERS viruses are known, this has yet to have been confirmed for SARS-CoV-2. All three strains are transmissible between humans.
To allow the widest possible distribution of relevant research, the Microbiology Society has brought together articles from across our portfolio and made this content freely available.
Image credit: "MERS-CoV" by NIAID is licensed under CC BY 2.0, this image has been modified.
-
-
-
CRISPR
More Less
The discovery of the CRISPR-Cas system has led to a sea of change in the microbiological sciences. Since then laboratories all over the world have joined the race to understand and exploit CRISPR. The insights gained about this system led to applications in industry to protect bacterial species against their viral parasites. In addition, CRISPR-Cas has been turned into a versatile genome editing method that has the potential to treat human genetic diseases.
Find out more about CRISPR in this collection of articles by watching this video.
Image credit: iStock/ibreakstock
-
-
-
David Rowlands collection
More Less
Each year, the Microbiology Society Council offer Honorary Membership to distinguished microbiologists who have made a significant contribution to the science. In 2019, David J. Rowlands (Emeritus Professor of Virology, University of Leeds) was appointed an Honorary Member.
This collection brings together Journal of General Virology articles authored by David Rowlands.
-
-
-
Diversity in Microbiology
More Less
As the President of the Microbiology Society, I am eager that we continue to build on our work supporting Equality, Diversity and Inclusion (EDI). Greater diversity within all that we do will widen the talent pool available for the field of microbiology and create networks of ideas and collaborations, potentially leading to greater development and innovation. Working to be inclusive helps us ensure we have a thriving community, which in turn will pave the way for us to support microbiology into the future. I have commissioned a number of articles from under-represented groups, highlighting the talent we have within our Society, and where our members are producing cutting-edge research within our discipline.
Professor Gurdyal Besra, Microbiology Society President
Photo credit: iStock/ Angelina Bambina
-
-
-
Ebola Virus Disease (EVD)
More Less
Ebola first became headline news in 2014–2016 when it was transmitted throughout West Africa. In 2019, this severe and often fatal disease has once again been declared a public health emergency of international concern (PHEIC) with over 1700 deaths in this latest outbreak. With vaccines now available, this outbreak could be contained, but only with increased production and delivery of vaccines within the Democratic Republic of Congo.
This collection brings together articles from our portfolio of journals on Ebola virus disease. The Microbiology Society has made this content freely available in the interests of widest possible distribution of relevant research.
-
-
-
Environmental Sensing and Cell-Cell Communication
More Less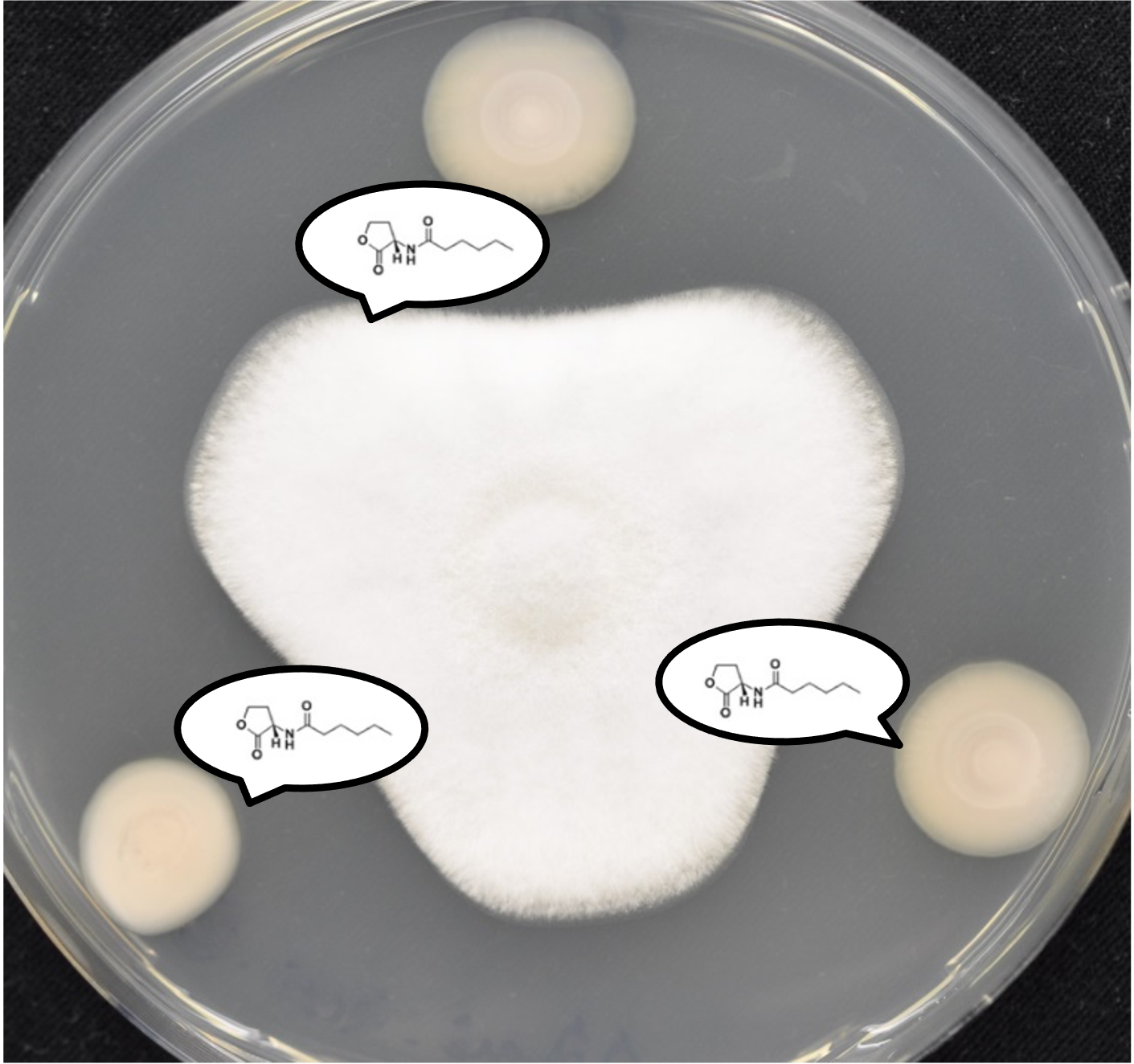
The last two decades have provided a wealth of new insight into how microbes (both prokaryotes and eukaryotes) sense and respond to their surroundings and to one another. Technological advances continue to shape our understanding of this burgeoning field, and this has led to a sea-change in the way in which we view the microbial world. No longer are microbes viewed as being the archetypal single celled entities; instead, community spirit and coordinated responses are the order of the day. In this special anniversary collection for Microbiology, timed to coincide with the Microbiology Society-sponsored Cell-Cell Communication meeting, Guest Editors Martin Welch (University of Cambridge) and Anugraha Mathew (University of Zurich) aim to assemble a landmark collection of papers that celebrate the interaction of microbes with their environment and with one another.
Submissions are particularly welcomed on microbial sensing and signaling pathways, quorum sensing (including both intra- and inter-species interactions and other forms of community-wide behaviors), chemoreception, secondary metabolism, and the complex interplay between different sensory pathways.
-
-
-
Establishing whole genome sequencing at the core of epidemiological surveillance
More Less
Over the last two decades, genome sequencing has become an important tool for understanding and tracking the spread of pathogens. Genomic epidemiology is now a preferred method of surveillance and recent years have seen pathogen sequencing at an unprecedented scale, pushing the underlying technologies to the limit. This has brought major innovations and opportunities to public attention, as well as identifying new research areas. However, major challenges remain in public health settings. These include: incorporating new sequencing technologies and data types for real-time surveillance; developing platforms and nomenclatures for genome-based typing and epidemiology; understanding pathogen evolution and the emergence of virulence and antimicrobial resistance; contextualizing knowledge of clinical microbiology with One Health ecological genomics. In this collection, we bring together recent studies that are establishing pathogen genomics as a major part of contemporary disease control efforts.
-
-
-
Ethnopharmacology
More Less
A concerted effort to characterise, assess and exploit the extensive written and oral record of natural products used in pre-modern European medicine has not been made. This is despite the presence in pre-modern European medical texts of natural products known to be effective in vivo (e.g. Artemisia spp. were used to treat malaria in medieval England) or shown to possess antimicrobial and/or immunomodulatory qualities in vitro (e.g. Allium spp., Plantago spp., Urtica spp.). Given rising antimicrobial resistance and a stalled R&D pipeline for compounds to treat and prevent infection, a thorough scientific evaluation of European ethnopharmacology is overdue.
This collection brings together original articles, mini-reviews, and full-length reviews written by researchers from diverse fields including microbiology, chemistry, botany and the history of medicine, along with industry contacts, to reveal the current state of the art of the field and define areas for collaboration, methods development and translational research.
-
-
-
Exploring the skin microbiome in health and disease
More LessBeing our biggest organ and a primary protective barrier from pathogens, the microbiome of this highly varied environment is now being described in unprecedented detail, revealing complex multispecies communities that play important roles in skin health, the development of the immune system and in wound healing. This collection aims to bring together knowledge of what these microbes are, how they colonise this often nutrient poor and challenging niche, and how they work together to suppress the growth of pathogens, while in themselves also being potential accidental pathogens if the skin barrier is broken.
This collection will feature new primary research and review articles arising from the “Exploring the skin microbiome in health and disease” symposium held at the Microbiology Society Annual Conference 2024 in Edinburgh, 8-11 April 2024.
The collection is also open for new submissions from all researchers across the skin microbiome field. Please indicate within your submission that it is intended for the collection.
Guest Editors: Georgios Efthimious (University of Hull, UK); Albert Bolhois (University of Bath, UK); Andrew Edwards (Imperial College London, UK)
Status: Open for submissions
Deadline for submissions: 7th October 2024
Journals submission links: Microbiology Editorial Manager
Journal of Medical Microbiology Editorial Manager
Microbial Genomics Editorial Manager
-
-
-
Feed the World
More Less
With the United Nations' resolution to eliminate world hunger by 2030 and the globe's population heading towards nine billion, the agriculture industry needs to increase livestock production from the same, or less, land. Livestock uses the most agricultural land (80% including grazing land and cropland for feed). Africa and Asia are the continents with the largest share of the world's uncultivated land, but attempts to develop and expand current capacity in order to meet the growing food demand are halted by deadly killers in the form of viruses, bacterial and protozoan parasites. As such, this area of research is hugely important to ensuring the availability of food for the world’s population.
The ‘Feed the World’ collection brings together articles published across the journal portfolio, focussing on food security, and agriculture and livestock diseases that have an economic impact on humans and animals. Guest edited by Alison Mather (Quadram Institute) and Nigel French (University of New Zealand) for Microbial Genomics; Colin Crump (University of Cambridge) for Journal of General Virology, and Sharon Brookes (Animal Plant Health Agency) for Journal of Medical Microbiology.
This collection is now accepting submissions via any participating journal. Please indicate that your submission is part of the ‘Feed the World’ collection.
-
-
-
Fungal spotlight: Host-associated microbiomes
More Less
Fungi comprise a distinct eukaryotic lineage often associated with their important role as degraders of organic substrates in the environment. However, fungi also form well-known symbiotic association with higher plants, as mycorrhizae, or with algae as lichens. Additionally, fungi are a critical component of the environmental microbiome associated with both plants and animals. The functional role of fungi in these interactions is poorly understood. Increasingly fungi are being recognized as opportunistic pathogens of animals, including humans, and as plant pathogens, are a threat to the global supply of food. Fungi themselves may harbour their own unique microbiome or organise the microbiome of the substrate they colonise.
This collection will feature studies of fungi in host-associated microbiomes, functional analysis of host-fungal or fungal-microbe interactions, the genetic and genomic diversity of host-associated fungi, as well as the impacts of environmental fungi in natural and manmade ecosystems. It is guest edited by Professor Corby Kistler (University of Minnesota), Dr. Ferry Hagen (Westerdijk Fungal Biodiversity Institute), Dr. David Fitzpatrick (Maynooth University), and Dr. Daniel Croll (University of Neuchâtel).
Image credit: Corby Kistler (University of Minnesota), US Department of Agriculture (public domain), and Biotec, Thailand
-
-
-
Genomics, Epidemiology and Evolution of Campylobacter, Helicobacter and Related Organisms
More Less
The Campylobacterales are typically microaerophilic species that are often host adapted with the potential for zoonotic infections. The order includes Campylobacter which is the most common bacterial source of gastroenteritis worldwide; Helicobacter which have been associated with peptic ulcers, chronic gastritis, duodenitis, and stomach cancer; the veterinary and clinical pathogen Arcobacter and other related organisms. In conjunction with recent conferences, we present a collection of contemporary studies investigating the genomics of these organisms, guest-edited by members of the conference organising committees: Dr Beile Gao (CHRO-2022; Chinese Academy of Sciences); Dr Ben Pascoe (CampyUK-2021; University of Oxford) and Professor Sam Sheppard (Ineos Oxford Institute, University of Oxford). The Genomics, epidemiology and evolution of Campylobacter, Helicobacter and Related Organisms collection will bring together recent advances that use genomic data to advance understanding in the field. Wide-ranging contributions include findings from active national surveillance programs, comparisons of local and global variations in population structure, studies of antimicrobial resistance emergence and spread, core and accessory genome evolutionary analyses, pathogenicity studies, investigations of plasmids and mobile elements, and genome rearrangement, editing and/or methylation studies.
-
-
-
Global Pneumococcal Sequencing collection
More Less
Streptococcus pneumoniae is a leading cause of pneumonia, septicaemia and meningitis in young children. The global deployment of pneumococcal conjugate vaccine (PCV) which targets up to 13 of the >100 serotypes has effectively reduced the incidence of pneumococcal disease in children worldwide. However, there are still ~9 million cases of disease and over 300,000 deaths annually, in part due to replacement with non-vaccine serotypes and persistence of vaccine serotypes.
The Global Pneumococcal Sequencing Project (GPS) is a worldwide genomic surveillance network of >100 scientists. The project is set out to investigate the adaptive changes in pneumococcal populations before and after the vaccine introduction through whole-genome sequencing. At present, the project has sequenced >26,000 pneumococcal isolates from >50 countries and identified emerging lineages with multidrug resistant and high virulence comprising the benefit of PCV programme at international level.
This collection includes genomic surveillance analyses at the national-level and captures important insights, including identification of emerging serotypes and lineages after vaccine implementation. Such insights can inform vaccine use at the national level, and the design of future vaccines. Many of these studies were led by in-country partners, working closely with the GPS team, to enhance local capacity for future genomic surveillance of pathogens.
-
-
-
ICTV Virus Taxonomy Profiles
More Less
Journal of General Virology ICTV Virus Taxonomy Profiles are a freely available series of concise, review-type articles that provide overviews of the classification, structure and properties of individual virus orders, families and genera.
ICTV Virus Taxonomy Profiles are written by the International Committee on Taxonomy of Viruses (ICTV) study groups, comprised of leading experts in the field. The profiles summarise the individual chapters from the ICTV’s online 10th Report on Virus Taxonomy, and provide the latest taxonomic information on viruses.
The Microbiology Society is publishing these citable profiles online, while the full chapters are available to all through the ICTV website, thanks to a five-year Biomedical Resources grant from the Wellcome Trust.
-
-
-
Implementation of Genomics in Clinical and Public Health Microbiology
More Less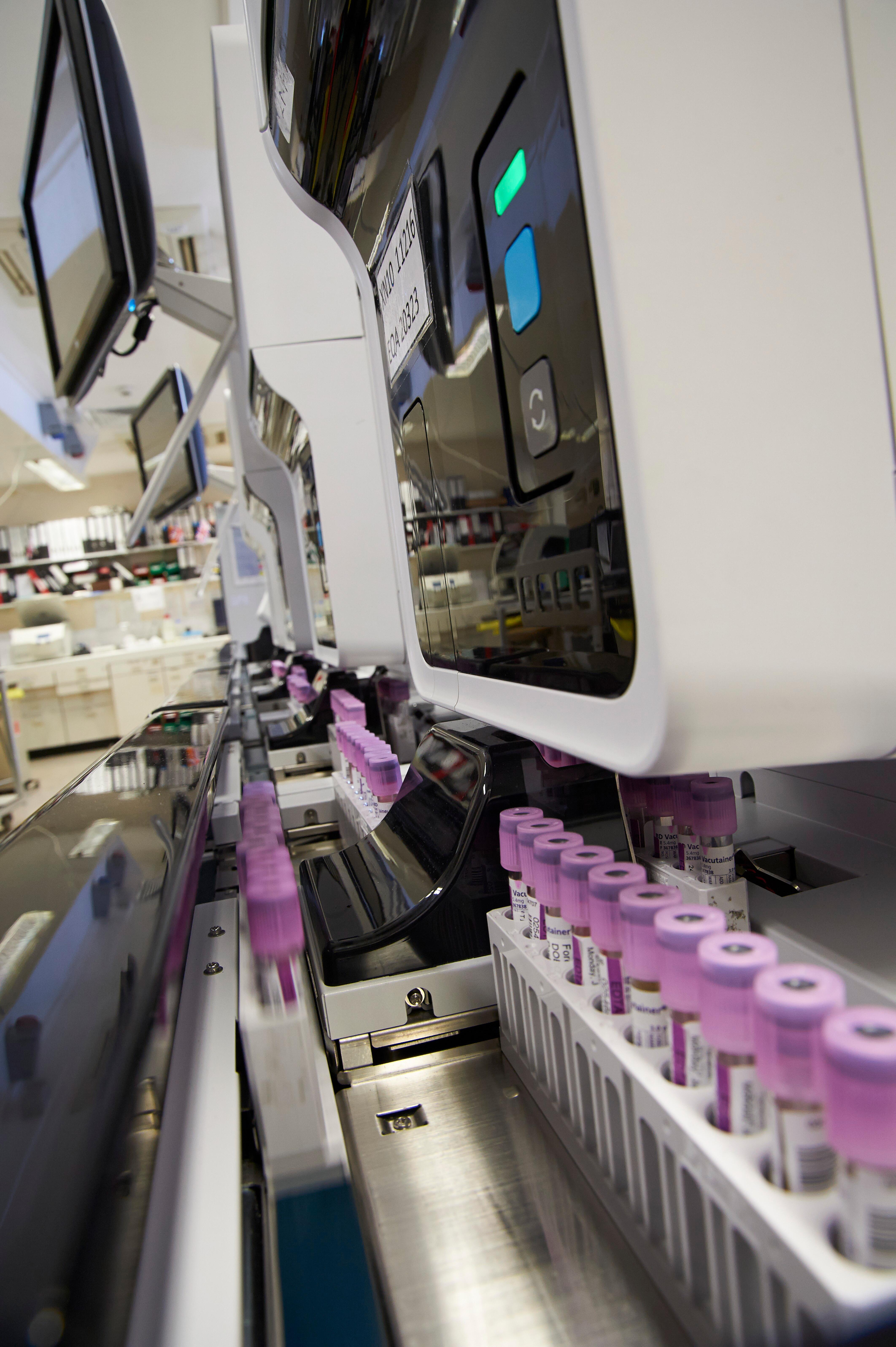
Microbial sequencing is rapidly being incorporated into public health and clinical microbiology, informed by over two decades of microbial genomics research and methods development. However, the integration of genomics into public health and clinical services and workflows is complex and requires consideration of a wide range of issues including workflow management, data analysis and reporting, and evaluation of how microbial genomic data can and should be used to support decision-making. This collection puts the spotlight on studies supporting microbial genomics implementation – aiming to understand what, why, and how microbial genomics works in “real world” clinical and health laboratory settings, and testing approaches to improving the integration of genomics into routine microbiological diagnostics, surveillance and outbreak investigation.
Image: Samples in a laboratory. Credit: Adrian Wressell, Heart of England NHS FT. Attribution 4.0 International (CC BY 4.0)
-
-
-
Implications of climate change for terrestrial microbiomes and global cycles
More Less
Soil microbiomes are highly diverse ecosystems that play a central role in ecosystem functioning and regulating global biogeochemical cycles. Through these processes, soil microbes make major contributions to the production and consumption of greenhouse gases. They also regulate the availability of essential nutrients, such as nitrogen and phosphorous, thereby influencing plant growth and global primary productivity. As such, soil microbial communities are intimately involved in climate feedback processes.
Predicting the contribution and response of soil microbiomes to future climate change represents a major research challenge. For example, uncertainties remain around the extent to which soils will act as a source or sink of carbon under future climate scenarios. A greater understanding of the microbial molecular pathways involved in biogeochemical cycling will be essential if we are to predict these outcomes. Similarly, further insight is needed into the response of soil microbial communities to climate extremes, such as drought, floods and increasing salinity, so that we can predict and mitigate changes to vital ecosystem services.
The special collection ‘Implications of climate change for terrestrial microbiomes and global cycles’ guest-edited by Drs Michael Macey (Open University), Sarah Worsley (UEA), and Geertje van Keulen (Swansea University), aims to highlight key research investigating the role of soil microbiomes in climate feedback processes, and their response to global change. It will also include articles on the characterisation of biogeochemical cycles and terrestrial microbiomes. We would also like to invite contributions on advances made in affordable and sustainable research methods, e.g. focussing on in situ activities where access to energy and/or data sources may be unreliable or unavailable.
This collection is open for submissions – please submit your article here, stating that your manuscript is part of the ‘Implications of climate change for terrestrial microbiomes and global cycles’ collection.
Image credit: Guido Gerding - external homepage, CC BY-SA 3.0, via Wikimedia Commons
-
-
-
International Coronavirus Networks Collection
More Less
US Food and Drug Administration (FDA), the UK International Coronavirus Network (UK-ICN) - Biotechnology and Biological Research Council (BBSRC) and the Department of Environment, Food and Rural Affairs (DEFRA), the Biosafety Level 4 Zoonotic Laboratory Network (BSL4ZNet), the Commonwealth Scientific and Industrial Research Organisation (CSIRO) collective bring together findings and lessons learnt for science foresight perspectives on Coronaviruses into our future.
The scope of the collection includes:
What we know: SARS, MERS, HCoVs and COVID-19
Emergence: Animal-human-animal interfaces
ABC: Bringing AIML, Bioinformatics and Coronavirology together
Social Contributions: Impacts of human and animal behaviours
Known Unknowns: Bat betacoronaviruses with Disease X potential
Prevention and Cure: Medical countermeasures (human and veterinary)
-
-
-
JMM Profiles
More Less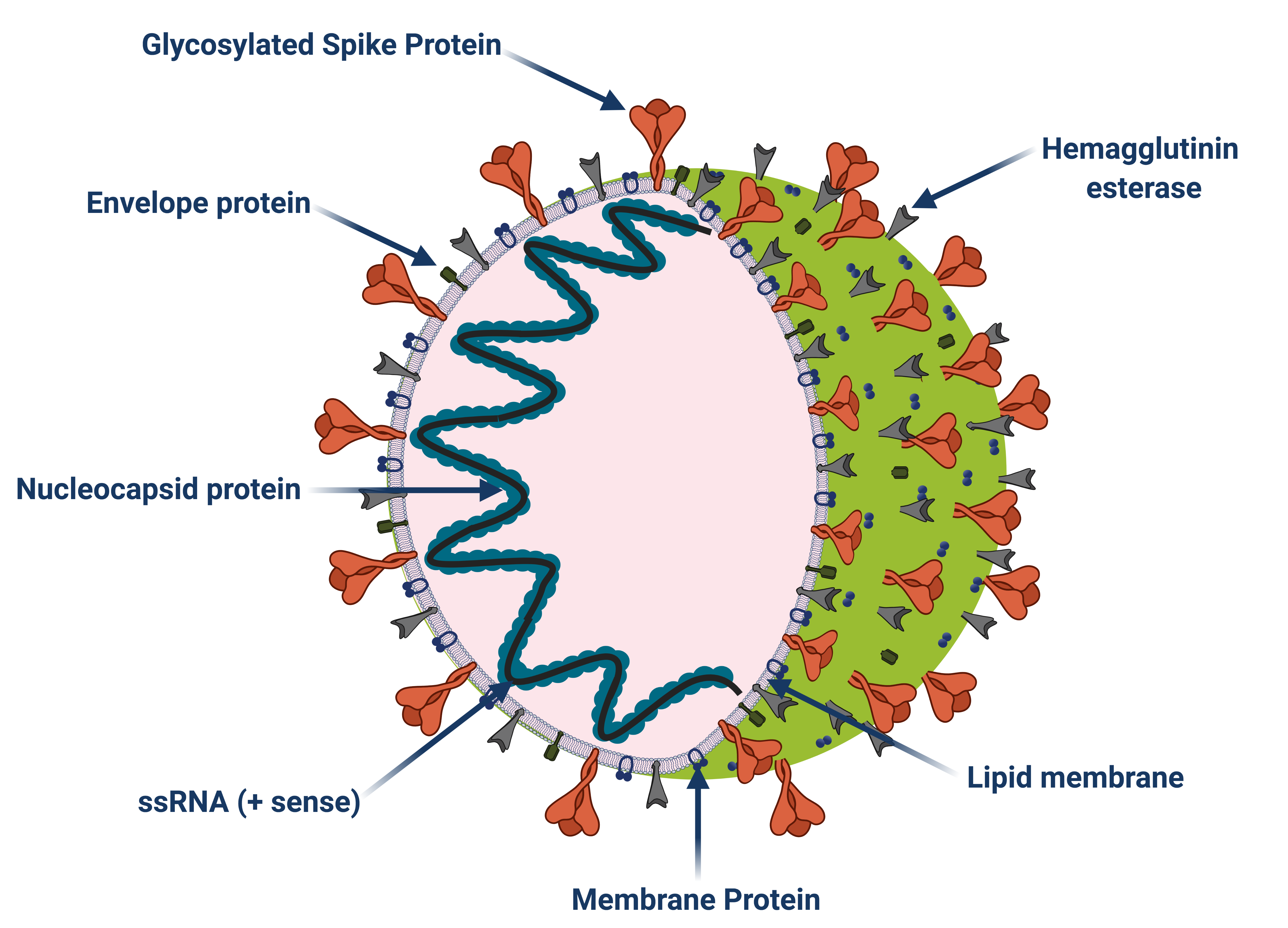
Journal of Medical Microbiology is now publishing ‘JMM Profiles’ in one of three categories; Pathogen Profile, Antimicrobial Profile and Diagnostic Profile. These articles will provide a brief summary review in each of these areas. The profiles are fully citable and free to read for 30 days after publication, and will make an excellent resource for education or reference.Please see our introductory Editorial for further information.
-
-
-
Knocking Out AMR
More Less
Antimicrobial resistance (AMR) is one of most urgent global threats to our healthcare systems, economies, the environment and animal health. Microbiologists in academic, industrial and clinical settings worldwide are at the forefront of developing innovative solutions to tackle AMR. We believe now is the time to act and focus on an interdisciplinary, solutions-driven approach in a ‘One Health’ context. To this end, the Microbiology Society is launching the ‘Knocking Out AMR’ project, an ambitious, bold and extensive scheme of work aiming to promote feasible and effective solutions to AMR through cross-disciplinary and multi-sector collaboration worldwide.
To accompany this, we are building a Knocking Out AMR collection to serve as a central hub for the AMR content published in our portfolio. This page will soon be updated with new content as well as relevant research already published in our portfolio. In the meantime, check out our existing AMR collections:
-
-
-
Marine Microbiology
More Less Covering over 70% of the Earth’s surface, the Oceans represent an incredibly diverse, yet understudied ecosystem. In particular, microorganisms (bacteria, fungi, viruses and archaea) mediate key ecosystem processes in these marine systems and account for a majority of the biodiversity. These resilient microbial species have evolved by partitioning a multitude of environmental resources including variable temperature, ocean currents, light availability, pressure and nutrients. At the same time, the dynamic nature of marine systems allows for complex biotic interactions that aggregate into microbiomes, or microbial communities. Various environmental factors influence biogeographic patterns that allow for distinct community assemblages across marine habitats (e.g., water column vs. sediment).
Covering over 70% of the Earth’s surface, the Oceans represent an incredibly diverse, yet understudied ecosystem. In particular, microorganisms (bacteria, fungi, viruses and archaea) mediate key ecosystem processes in these marine systems and account for a majority of the biodiversity. These resilient microbial species have evolved by partitioning a multitude of environmental resources including variable temperature, ocean currents, light availability, pressure and nutrients. At the same time, the dynamic nature of marine systems allows for complex biotic interactions that aggregate into microbiomes, or microbial communities. Various environmental factors influence biogeographic patterns that allow for distinct community assemblages across marine habitats (e.g., water column vs. sediment).
Ultimately, understanding the relationship between ecological and evolutionary processes and the environment will elucidate the factors driving marine microbial distributions and community structure. In order to understand these complex dynamics, marine microbiology necessitates interdisciplinary science, encompassing fields such as biogeochemistry, oceanography, ecology, geology, chemistry and microbiology. It also represents incredible potential for discovery, with impacts on our ability to manage and utilize the oceans as a sustainable resource.
Guest-edited by Dr. Katherine Duncan and Dr. Alex Chase, this Marine Microbiology special collection aims to highlight key research on marine microorganisms as they underpin the complex processes of our blue planet.
-
-
-
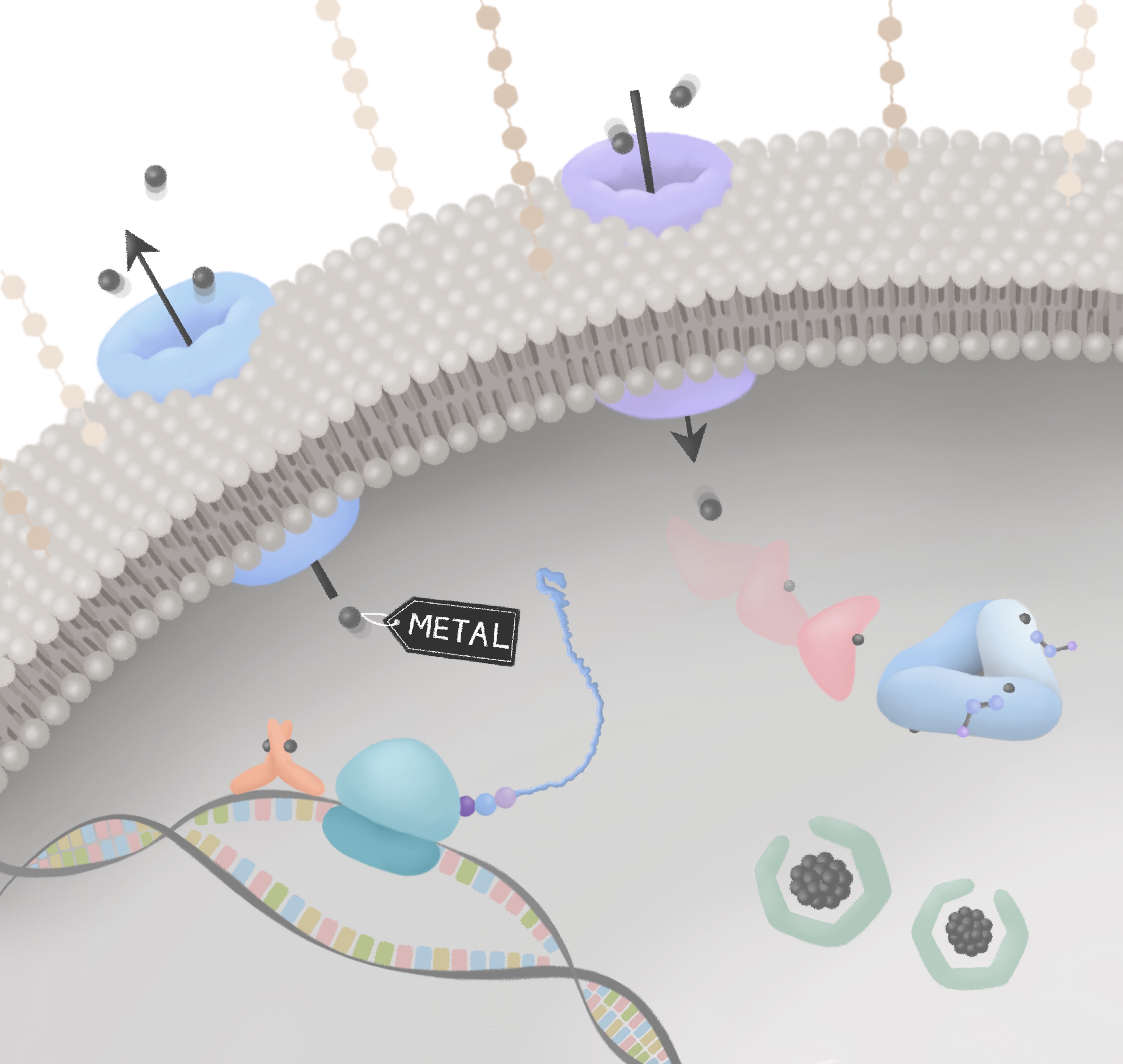
Metals in Microbiology
More Less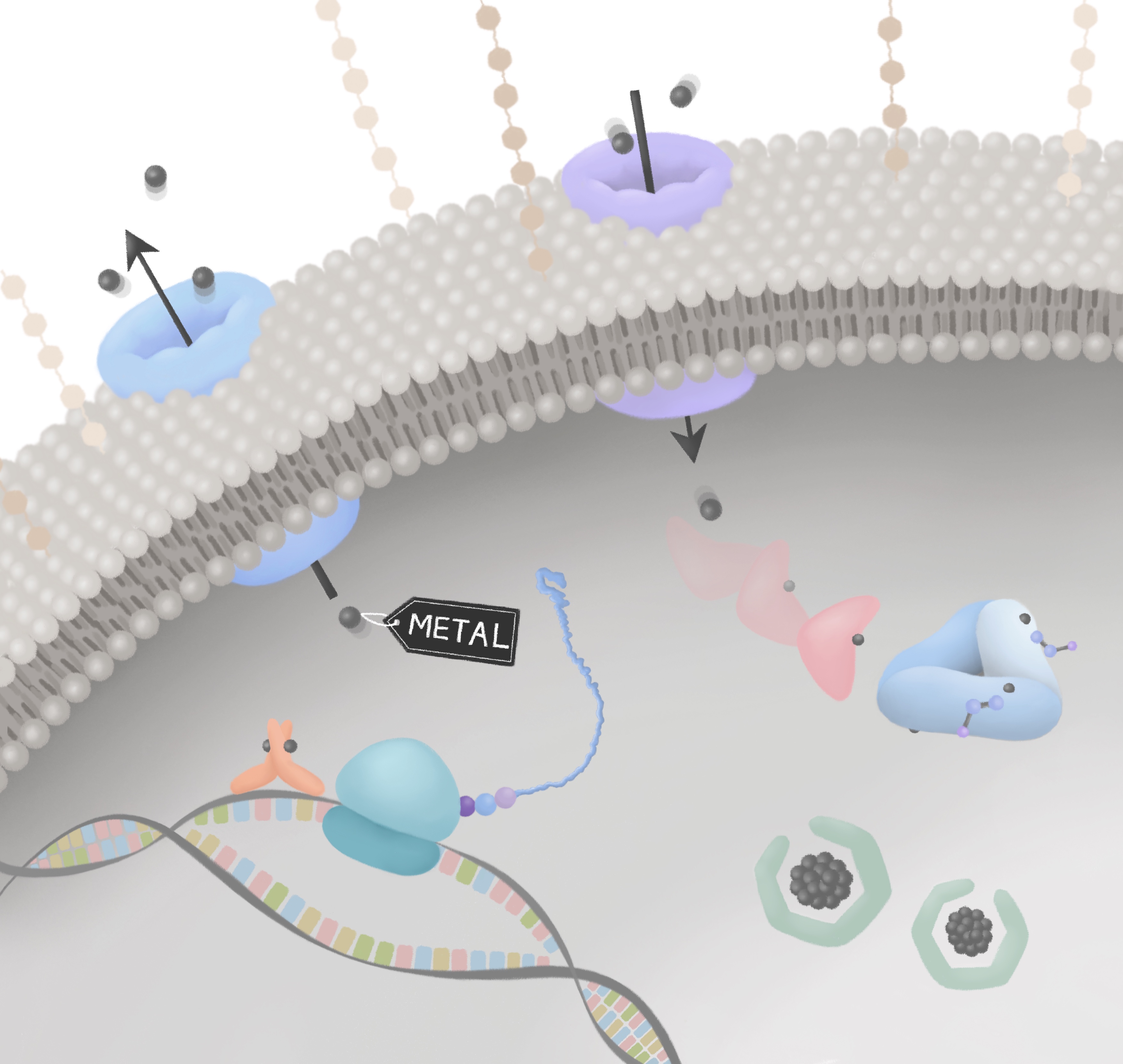
Metals catalyse almost a half of all microbial reactions and yet can poison microorganisms. Metal handling systems - that maintain metal homeostasis - are thus vital to sustain microbial life. For microbial pathogens, the challenge of metal homeostasis is exacerbated by host immune defences that restrict metal access and that exploit the microbicidal activity of metals. There are opportunities to produce new antimicrobials that subvert microbial metal-handling systems or that use metals directly or combined with other compounds. The prevalence of metalloenzymes also means that engineering the metal-supply in microorganisms is highly relevant to industrial biotechnological processes, with metalloproteins contributing to bioenergy production, bioremediation, biomedicine, synthesis of high value industrial feedstocks and more.
Guest-edited by Dr Jennifer Cavet (University of Manchester) and Dr Karrera Djoko (Durham University), this collection of keynote research articles will highlight research on metal-microbe interactions, bringing together advances in our understanding of how microbes handle metals, the utilization of metals in proteins and the importance of metal handling systems in host-pathogen interactions. It will also include research that exploits these systems in industrial processes, the development of metal-related antimicrobials and in metal bioremediation and biorecovery.
Image credit: Jin Hong
-
-
-
Methods and Software
More Less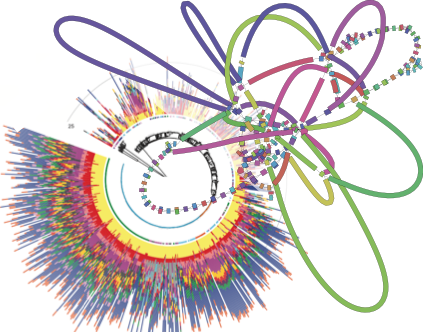
The Microbial Genomics Methods and Software collection will bring together articles describing novel experimental, bioinformatics, modelling, and statistical approaches to the analysis of microbial genomics data, including databases or the integration of genomics with other data streams; as well as systematic comparisons or benchmarking of existing methodologies used in the field of microbial genomics. Guest-edited by Dr Zamin Iqbal (European Bioinformatics Institute) and Dr Caroline Colijn (Simon Fraser University), the collection aims to provide the microbial genomics community with new and systematically validated tools to advance their research.
The cover image for this collection brings together figures from two of retrospective articles in the collection: a phylogeny richly annotated with insertion sequence sites from the article on ISseeker by Adams et al. 2016 (bottom left); and a genome assembly graph from the article on completing bacterial genomes by Wick et al. 2017 (top right).
This collection is now open for submissions. Submit your article here, stating that your manuscript is part of the Methods and Software collection.
-
-
-
Micro Perspectives
More Less
Our community provide opinions, views and authoritative perspectives on work published in our journals.
-
-
-
Microbe Profiles
More Less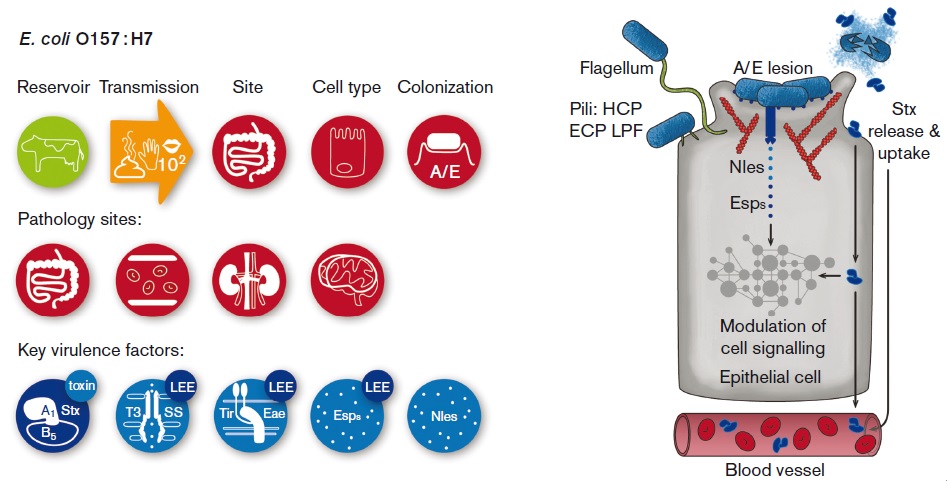
Microbiology is now publishing ‘Microbe Profiles’ – concise, review-type articles that provide overviews of the classification, structure and properties of novel microbes, written by leading microbiologists. These profiles will provide insights into key microbes within the field. The profiles are fully citable and free to read for 30 days after publication, and will make an excellent resource for education or reference.
-
-
-
Microbial Evolution
More Less Microbes are everywhere, living inside and around us, often within complex and dynamic communities that underpin the health of our bodies and of natural ecosystems. The need to understand and predict microbial evolutionary dynamics has never been more urgent. The rise of antimicrobial resistance is a crisis caused by the evolutionary adaptation of microbes to our use of antibiotics. How microbes respond to global change will shape critical biogeochemical processes in oceans and soils. Solutions to these and many other emerging issues will require an in-depth understanding of the evolutionary dynamics of microbial communities, to enable us to predict and manage their responses to selective pressures and to design robust biotechnological solutions. This collection will highlight microbial evolution research papers from the Microbiology archives and feature new primary research and review articles arising from the “Understanding and Predicting Microbial Evolutionary Dynamics” Focus Meeting held in Manchester 22-23 November 2022.
Microbes are everywhere, living inside and around us, often within complex and dynamic communities that underpin the health of our bodies and of natural ecosystems. The need to understand and predict microbial evolutionary dynamics has never been more urgent. The rise of antimicrobial resistance is a crisis caused by the evolutionary adaptation of microbes to our use of antibiotics. How microbes respond to global change will shape critical biogeochemical processes in oceans and soils. Solutions to these and many other emerging issues will require an in-depth understanding of the evolutionary dynamics of microbial communities, to enable us to predict and manage their responses to selective pressures and to design robust biotechnological solutions. This collection will highlight microbial evolution research papers from the Microbiology archives and feature new primary research and review articles arising from the “Understanding and Predicting Microbial Evolutionary Dynamics” Focus Meeting held in Manchester 22-23 November 2022.
This collection is open for new submissions from all researchers across the full breadth of the microbial evolution field and is guest edited by Michael Brockhurst (University of Manchester, UK), Jenna Gallie (Max Planck Institute for Evolutionary Biology, Germany), James Hall (University of Liverpool, UK), and Stineke Van Houte (University of Exeter, UK).The collection will be extended to also encompass new articles arising from Microbial Ecology and Evolution Hub-based Conference 2024 (MEEhubs2024), and submissions related to this meeting will be guest edited by Wolfram Moebius (University of Exeter, UK).Image credit: Science Photo Library/KuLouKu
-
-
-
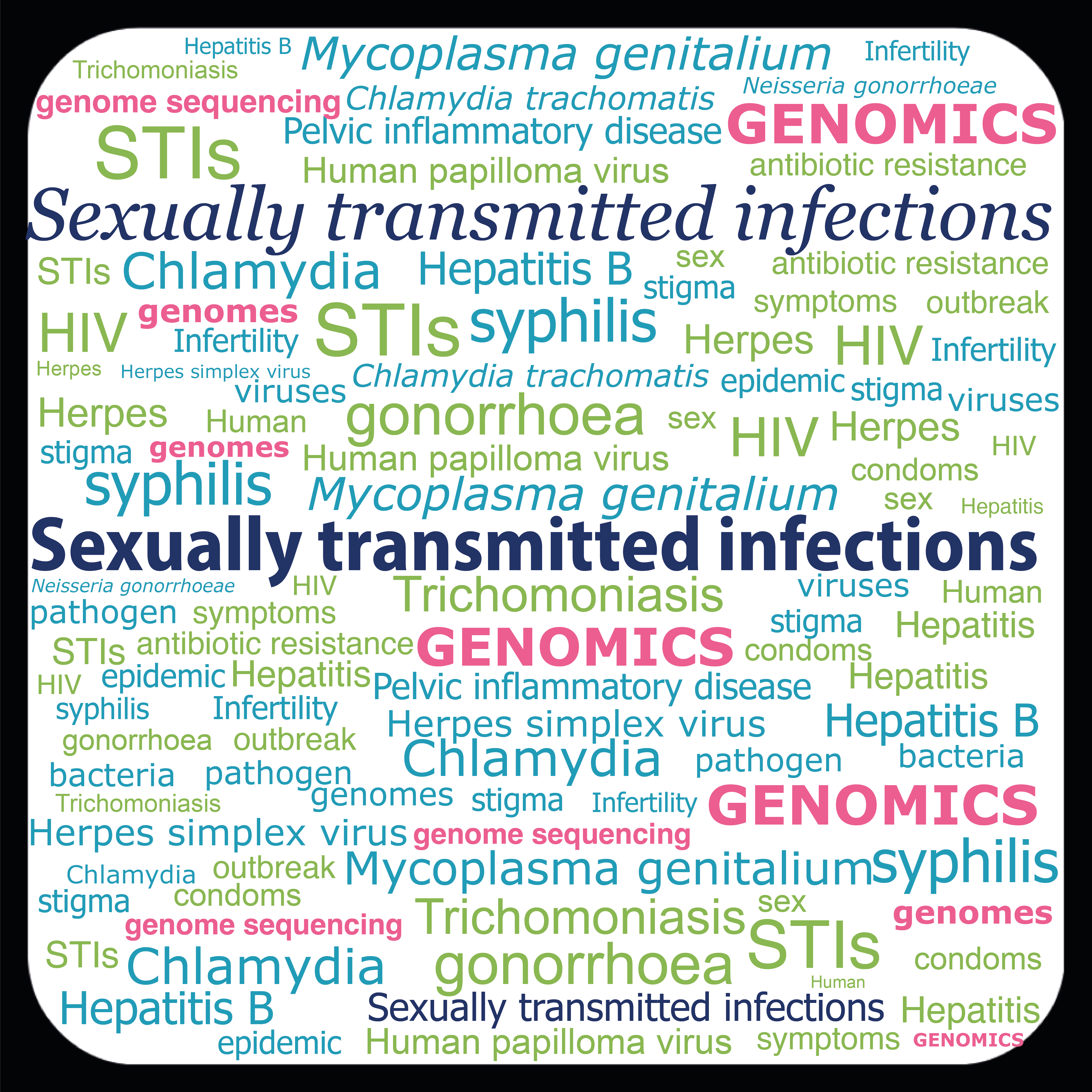
Microbial Genomics of Sexually Transmitted Infections
More Less
Sexually transmitted infections (STIs) remain a major global public health threat. Often disproportionately impacting specific groups of people, significant morbidity and mortality is caused by common diseases including gonorrhoea, chlamydia, syphilis, HIV infection, Hepatitis B, and disease linked to Human Papilloma Virus. STI Control can be difficult and challenges range from practical obstacles around the distribution of effective antivirals and vaccines to the emergence of antimicrobial resistance (AMR) among bacterial pathogens. Undoubtedly, effective interventions depend upon detailed understanding of the pathogens and whole genome sequencing (WGS) provides a solution to this. With improved access to resources and decreasing sequencing costs, WGS has dramatically changed the research landscape revealing the genetic basis of important phenotypes such as virulence and AMR, underpinning local and national surveillance programs to understand disease epidemiology, emergence and spread, and accelerating vaccine development.Guest-edited by Dr Odile Harrison and Dr Janet Wilson (President of IUSTI) this collection gathers original research articles, reviews and methods that use WGS approaches for the analysis of sexually transmitted pathogens. The goal of this collection is to provide to the community a comprehensive suite of articles that consolidate recent advances made in the field and provide tools for the community for further research.
This collection is now open for submissions in Microbial Genomics. If you’d be interested in being included in the collection, please submit to our peer review system and state that your submission is intended for the ‘Microbial Genomics of Sexually Transmitted Infections’ collection.
Image credit: Odile Harrison
-
-
-
Microbial Primers
More Less
In the constantly evolving world of microbiology, new research and discoveries can often be overwhelming. ‘Microbial Primers’ is a series of short articles designed to simplify and illuminate intricate microbiological concepts, making them easily digestible and accessible. Each article in the series focuses on a specific subject, distilling it to the most fundamental points, and explaining its importance in the wider scientific context. The series serves as a starting point, to encourage further exploration and discovery in the fascinating world of microbes. Whether you're an early career researcher or an established scientist looking to broaden your horizons, ‘Microbial Primers’ is your first-step navigational tool in the expansive landscape of microbiology.
Photo credit: iStock/SpicyTruffel
-
-
-
Unlocking the world of microbiomes
More Less
In 2020 we celebrate 75 years of the anniversary of our founding with a year of activities dedicated to demonstrating the impact of microbiologists’ past, present and future – bringing together and empowering communities that help shape the future of microbiology. We are launching new collections of digital content throughout the anniversary year. The first digital hub is Unlocking the world of microbiomes: exploring microbial communities, which will examine the microbiome and human health, agriculture and food microbiomes and environmental and industrial microbiomes.
The ‘Unlocking the world of microbiomes’ collection brings together articles from across our journals exploring microbial communities and examining the microbiome and human health. This collection is an update of a collection by the Microbiology Society and the British Society for Immunology launched for World Microbiome Day; the ‘Microbiome’ collection can be viewed on Science Open.
-
-
-
Microreact
More Less
Microbial Genomics is partnered with Microreact, a free data visualisation and sharing platform that allows scientists and health professionals worldwide to better collaborate to understand disease outbreaks. The journal encourages authors to upload their data files to Microreact, which can then provide interactive querying of the data via trees, maps, timelines and tables, and will be published in Microbial Genomics via a permanent web link. This collection brings together articles that have data sets generated by this software.
If you would like to see your work included in this collection, please click here for more information, or submit your article here.
-
-
-
Most Downloaded Articles of 2018
More Less
We are delighted to present a collection of the Microbiology Society’s most downloaded journal content from January–September 2018. This collection highlights the exciting and impactful research across the breadth of microbiology and is an easy way for you to quickly access the content your peers are reading.
-
-
-
Mycobacteria
More Less
Mycobacteria are a vast group of microorganisms characterized by a unique thick, hydrophobic cell wall rich in mycolic acids, which makes them highly resistant to environmental stresses. Even if most of them are innocuous environmental saprophytes, some of them, such as Mycobacterium leprae or Mycobacterium tuberculosis, have evolved to become formidable human pathogens with a very complex and still not well-characterized relationship with their host, while others, such as Mycobacterium avium, represent important emerging or opportunistic pathogens.
Guest-edited by Dr. Riccardo Manganelli, this collection of keynote research articles will highlight all aspects of mycobacterial biology, with particular focus on physiological aspects, such as stress response mechanisms, regulatory networks, and metabolic pathways, that might lead to a better understanding of the intriguing aspects of mycobacterial host-pathogen interaction and lead to the design of new strategies to fight these important pathogens.
-
-
-
Negative Results
More Less
Access Microbiology provides a platform to publish sound science across the entire field of microbiology. Negative results are an integral part of sound science and research integrity, adding value to the scientific literature that is often overlooked. In line with the platform’s mission, we have collated impactful negative results studies published since Access Microbiology’s launch in 2019. Science integrity consultant Elisabeth Bik provides an editorial for this collection, on the importance and value of negative data.
With ongoing debate in the scientific community about research integrity and the pressure to publish novel results, we want to highlight the high quality negative results studies published on the platform. New papers will be added to the collection as these are published.
-
-
-
New Frontiers in Microbiology
More Less
In 2020 we celebrate 75 years of the anniversary of our founding with a year of activities dedicated to demonstrating the impact of microbiologists past, present and future – bringing together and empowering communities that help shape the future of microbiology. We are launching new collections of digital content throughout the anniversary year.
As we have progressed through the 21st century, we have expanded and developed our understanding of how microbes are related to and interact with each other. Microbiology research has been, and continues to be, central to meeting many of the current global aspirations and challenges, such as maintaining food, water and energy security for a healthy population on a habitable earth. The ‘New Frontiers in Microbiology’ collection brings together articles on the ever-growing tree of life and synthetic biology.
-
-
-
Outbreak Reports
More Less
This collection highlights Outbreak Reports published in Microbial Genomics. Outbreak reports are short-format articles that investigate the key role that genomics plays in investigating communicable disease outbreaks.
If you would like to see your work included in this collection click here for more information. Microbial Genomics is looking for novel and interesting stories describing unique applications of genomics at any stage of an outbreak.
-
-
-
Pedagogy
More Less
Innovative teaching is now considered a crucial factor when designing and delivering high quality curricula and includes activities such as games, teamwork workshops and problem-solving sessions. In addition, higher education institutions are now expected to build strong bonds with the public via various outreach activities with schools or other community groups. In order to help modern microbiology educators stand up to these challenges, Access Microbiology provides a collection of pedagogical research in the field of microbiology, offering a handy database with all pedagogical papers published in Access Microbiology since 2019, updated regularly as new articles are published. This collection aims to help microbiology educators improve their teaching by introducing new innovative teaching and outreach ideas to their curricula, often generated by members and friends of the Microbiology Society.
-
-
-
Prokaryotic Stress Responses – their diversity and regulation
More LessMicroorganisms encounter a wide range of stresses and environmental changes in diverse scenarios including infection, ecological and biotechnological scenarios. By definition, stress is a driver of diversity, evolution and phenotypic heterogeneity. There are a wide range of prokaryotic stress responses, including antibiotic, envelope, host-derived, metabolic, starvation, environmental, redox, temperature, solvent and DNA damage stress responses. Microbes are also able to sense chemical stress, either via sensors at the cell surface or by cytoplasmic transcriptional regulators. Systems that have been identified in stress response research have been exploited for new treatments or increased productivity are welcomed. This collection aims to celebrate the responses induced by various stresses on diverse microbial taxa and the enabling technologies allowing their investigation including transcriptomics, metabolomics, biophysics and imaging.
This collection will feature new primary research and review articles arising from the “Prokaryotic Stress Responses – their diversity and regulation” symposium held at the Microbiology Society Annual Conference 2024 in Edinburgh, 8-11 April 2024.
The collection is also open for new submissions from all researchers across the prokaryotic stress responses field. Please indicate within your submission that it is intended for the collection.
Guest Editors: Nick Tucker (University of Suffolk, UK); Dany Beste (University of Surrey, UK)
Status: Open for submissions
Deadline for submissions: 7th October 2024
Journal submissions link: Microbiology Editorial Manager
-
-
-
Pseudomonas
More Less
This collection brings together original research articles, mini-reviews, and full-length reviews relating to Pseudomonas. Guest edited by Dr Joanna Goldberg and co-edited by Dr Kalai Mathee, this collection will not only be relevant to scientists with an interest in Pseudomonas, but also, due to the widespread use of this genus as a model for studying multiple systems, it will be of general interest to other researchers active in areas such as evolutionary biology, communication systems, genomics and biofilm research. In addition, because P. aeruginosa is a key pathogen associated with both acute and chronic infections, and particularly important in the context of cystic fibrosis and antimicrobial resistance, the collection will be of interest to clinicians and clinical researchers.
This collection was released in conjunction with Pseudomonas 2019, an international conference in Kuala Lumpur, Malaysia.
Manuscripts submitted to the journal are free to publish. The Microbiology Society offers a Gold Open Access option, please see https://www.microbiologyresearch.org/publishing-costs for further details.
-
-
-
Streptomyces
More Less
Over the last century, Streptomyces bacteria – and their metabolic products – have revolutionized modern medicine. These little pharmaceutical factories produce a vast array of natural products that have been co-opted for medical and agricultural therapies. In addition to their metabolic sophistication, Streptomyces also exhibit remarkable developmental and regulatory complexity.
Guest-edited by Dr Marie Elliot, this collection of keynote research articles will highlight fascinating aspects of Streptomyces biology, and the advances that are providing us with newfound insight and appreciation for these extraordinary bacteria.
-
-
-
A Sustainable Future
More Less
To highlight the vital role microbiology plays in delivering on the UN Sustainable Development Goals (SDGs), we have created a collection of must-read research on three critical aspects of the SDGs: antimicrobial resistance, soil health, and the circular economy.
-
-
-
Symbiosis
More Less
Symbiosis has played a key role in the evolution of life on Earth. Symbiotic mergers of once independent species drove the origin of eukaryotes. Moreover, symbiosis has enabled many species to gain novel functions and occupy new ecological niches, thus underpinning the functioning of diverse ecosystems. As endosymbionts, microbes provide their eukaryotic hosts with an array of ecological and physiological innovations, including new metabolic capabilities, such as autotrophy or nitrogen fixation, and protection against infections or environmental stressors. Microbial eukaryotes also commonly host their own endosymbionts, including bacteria and algae. Understanding the stability and resilience of symbioses is key to predicting the response of important ecosystems, such as coral reefs, to global change. Manipulating symbiotic associations also has far-reaching economic, environmental and medical implications, through the potential to improve crop productivity, reduce reliance on fertilisers, and control the insect vectors of infectious diseases.
This collection, guest edited by Professor Michael Brockhurst (University of Manchester) and Dr. Rebecca J Hall (University of Birmingham), will feature microbe-focused studies of symbiosis, ranging from the molecular mechanisms of host-symbiont interactions, their genetic and genomic diversity, to understanding the impacts of symbioses in natural and manmade ecosystems.
-
-
-
Mycobacterium tuberculosis
More Less
World Tuberculosis Day on 24 March recognises the date in 1882 when Dr Robert Koch announced his discovery of Mycobacterium tuberculosis, the bacillus that causes tuberculosis (TB). In celebration of this, we are excited to present a collection of recently published papers on M. tuberculosis.
-











.jpeg)