
Full text loading...
Transmission Election Microscopy (TEM) image of Cellvibrio japonicus from a Hitachi HT7800 set to 100kV and stained with NanoW. The scalebar depicts 1µm. The polysaccharide diagrams shown are examples of substrates that the bacterium is adept at degrading. C. japonicus possesses an impressive array of carbohydrate active enzymes to cleave the glycosidic bonds of many plant, animal, and fungal polysaccharides.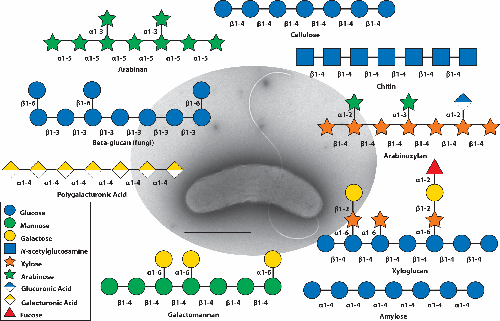
Cellvibrio japonicus is a saprophytic bacterium proficient at environmental polysaccharide degradation for carbon and energy acquisition. Genetic, enzymatic, and structural characterization of C. japonicus carbohydrate active enzymes, specifically those that degrade plant and animal-derived polysaccharides, demonstrated that this bacterium is a carbohydrate-bioconversion specialist. Structural analyses of these enzymes identified highly specialized carbohydrate binding modules that facilitate activity. Steady progress has been made in developing genetic tools for C. japonicus to better understand the function and regulation of the polysaccharide-degrading enzymes it possesses, as well as to develop it as a biotechnology platform to produce renewable fuels and chemicals.