
Full text loading...

Virulome profile of four Staphylococcus epidermidis strains isolated from platelet concentrates and human skin.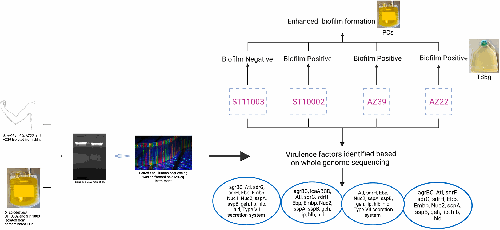
Staphylococcus epidermidis is one of the predominant bacterial contaminants in platelet concentrates (PCs), a blood component used to treat bleeding disorders. PCs are a unique niche that triggers biofilm formation, the main pathomechanism of S. epidermidis infections. We performed whole genome sequencing of four S. epidermidis strains isolated from skin of healthy human volunteers (AZ22 and AZ39) and contaminated PCs (ST10002 and ST11003) to unravel phylogenetic relationships and decipher virulence mechanisms compared to 24 complete S. epidermidis genomes in GenBank. AZ39 and ST11003 formed a separate unique lineage with strains 14.1 .R1 and SE95, while AZ22 formed a cluster with 1457 and ST10002 closely grouped with FDAAGOS_161. The four isolates were assigned to sequence types ST1175, ST1174, ST73 and ST16, respectively. All four genomes exhibited biofilm-associated genes ebh, ebp, sdrG, sdrH and atl. Additionally, AZ22 had sdrF and aap, whereas ST10002 had aap and icaABCDR. Notably, AZ39 possesses truncated ebh and sdrG and harbours a toxin-encoding gene. All isolates carry multiple antibiotic resistance genes conferring resistance to fosfomycin (fosB), β-lactams (blaZ) and fluoroquinolones (norA). This study reveales a unique lineage for S. epidermidis and provides insight into the genetic basis of virulence and antibiotic resistance in transfusion-associated S. epidermidis strains.

Article metrics loading...

Full text loading...
References


Data & Media loading...
Supplements
