
Full text loading...
 , Erin D. Gonzales2, Daniel B. Toso2,†, Natalie A. Kirk2,‡ and William J. Hickey2
, Erin D. Gonzales2, Daniel B. Toso2,†, Natalie A. Kirk2,‡ and William J. Hickey2
Stylized cryo-electron image of two bacterial cells and an extracellular vesicle highlighting the presence of a surface layer (gold-colored structure).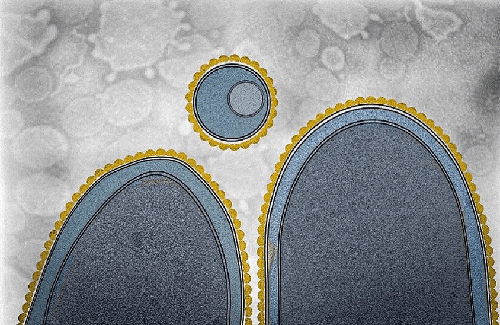
The phytopathogen Paracidovorax citrulli possesses an ortholog of a newly identified surface layer protein (SLP) termed NpdA but has not been reported to produce a surface layer (S-layer). This study had two objectives. First, to determine if P. citrulli formed an NpdA-based S-layer and, if so, assess the effects of S-layer formation on virulence, production of nanostructures termed nanopods, and other phenotypes. Second, to establish the distribution of npdA orthologs throughout the Pseudomonadota and examine selected candidate cultures for physical evidence of S-layer formation. Formation of an NpdA-based S-layer by P. citrulli AAC00-1 was confirmed by gene deletion mutagenesis (ΔnpdA), proteomics, and cryo-electron microscopy. There were no significant differences between the wild-type and mutant in virulence assays with detached watermelon fruit. Nanopods contiguous with S-layers of multiple biofilm cells were visualized by transmission electron microscopy. Orthologs of npdA were identified in 62 Betaproteobacteria species and 49 Gammaproteobacteria species. In phylogenetic analyses, NpdA orthologs largely segregated into distinct groups. Cryo-electron microscopy imaging revealed an NpdA-like S-layer in all but one of the 16 additional cultures examined. We conclude that NpdA represents a new family of SLP, forming an S-layer in P. citrulli and other Pseudomonadota. While the S-layer did not contribute to virulence in watermelon fruit, a potential role of the P. citrulli S-layer in another dimension of pathogenesis cannot be ruled out. Lastly, formation of cell-bridging nanopods in biofilms is a new property of S-layers; it remains to be determined if nanopods can mediate intercellular movement of materials.

Article metrics loading...

Full text loading...
References


Data & Media loading...
Supplements
