 -
Volume 151,
Issue 10,
2005
-
Volume 151,
Issue 10,
2005
Volume 151, Issue 10, 2005
- Genes And Genomes
-
-
-
Intragenomic recombination in the highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans
More LessThe highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans is strongly associated with aggressive periodontitis in adolescents of African descent. DNA fingerprinting using the frequently cutting restriction enzyme MspI and multilocus sequence typing (MLST) showed that five strains of this clone were genetically virtually identical, although ribotyping of the six rrn genes and EcoRI RFLP analysis of the seven IS150-like elements revealed differences. PCR analyses demonstrated that these multi-copy sequences are subject to intragenomic homologous recombination, resulting in translocations or large inversions. The genome rearrangements were reflected in differences among 25 strains representing the JP2 clone in DNA fingerprinting using the rare-cutting restriction enzyme XhoI and resolved by PFGE. XhoI DNA fingerprinting provides a tool for studying local epidemiology, including transmission of this particularly pathogenic clone of A. actinomycetemcomitans.
-
-
- Pathogens And Pathogenicity
-
-
-
σ B contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB
More LessThe ability of Listeria monocytogenes to invade non-phagocytic cells is important for development of a systemic listeriosis infection. The authors previously reported that a L. monocytogenes ΔsigB strain is defective in invasion into human intestinal epithelial cells, in part, due to decreased expression of a major invasion gene, inlA. To characterize additional invasion mechanisms under the control of σ B, mutants were generated carrying combinations of in-frame deletions in inlA, inlB and sigB. Quantitative assessment of bacterial invasion into the human enterocyte Caco-2 and hepatocyte HepG-2 cell lines demonstrated that σ B contributes to both InlA and InlB-mediated invasion of L. monocytogenes. Previous identification of the σ B-dependent P2 prfA promoter upstream of the major virulence gene regulator, positive regulatory factor A (PrfA), suggested that the contributions of σ B to expression of various virulence genes, including inlA, could be at least partially mediated through PrfA. To test this hypothesis, relative invasion capabilities of ΔsigB and ΔprfA strains were compared. Exponential-phase cells of the ΔsigB and ΔprfA strains were similarly defective at invasion; however, stationary-phase ΔsigB cells were significantly less invasive than stationary-phase ΔprfA cells, suggesting that the contributions of σ B to invasion extend beyond those mediated through PrfA in stationary-phase L. monocytogenes. TaqMan quantitative reverse-transcriptase PCRs further demonstrated that expression of inlA and inlB was greatly increased in a σ B-dependent manner in stationary-phase L. monocytogenes. Together, results from this study provide strong biological evidence of a critical role for σ B in L. monocytogenes invasion into non-phagocytic cells, primarily mediated through control of inlA and inlB expression.
-
-
-
-
Production of ammonium by Helicobacter pylori mediates occludin processing and disruption of tight junctions in Caco-2 cells
More LessTight junctions, paracellular permeability barriers that define epithelial cell polarity, play an essential role in transepithelial transport, cell–cell adhesion and lymphocyte transmigration. They are also important for the maintenance of innate immune defence and intestinal antigen uptake. Ammonium (
 ) is elevated in the gastric aspirates of Helicobacter pylori-infected patients and has been implicated in the disruption of tight-junction functional integrity and the induction of gastric mucosal damage during H. pylori infection. The precise mechanism of the effect of ammonium and the molecular targets of ammonium in host tissue are not yet identified. To study the effects of ammonium on epithelial tight junctions, the human colon carcinoma cell line Caco-2 was cultured on permeable supports and the transepithelial resistance (TER) was measured at different time intervals following exposure to ammonium salts or H. pylori-derived ammonium. A biphasic response to treatment with ammonium was found. Acute exposure to ammonium salts or NH3/
) is elevated in the gastric aspirates of Helicobacter pylori-infected patients and has been implicated in the disruption of tight-junction functional integrity and the induction of gastric mucosal damage during H. pylori infection. The precise mechanism of the effect of ammonium and the molecular targets of ammonium in host tissue are not yet identified. To study the effects of ammonium on epithelial tight junctions, the human colon carcinoma cell line Caco-2 was cultured on permeable supports and the transepithelial resistance (TER) was measured at different time intervals following exposure to ammonium salts or H. pylori-derived ammonium. A biphasic response to treatment with ammonium was found. Acute exposure to ammonium salts or NH3/ derived from urea metabolism by wild-type H. pylori resulted in a 20–30 % decrease in TER. After 24 h, the NH4Cl-treated cells showed a partial recovery of TER. In contrast, the control culture, or cultures that were exposed to supernatants derived from urease-deficient H. pylori, showed no significant decrease in TER. Occludin-specific immunoblots revealed the expression of a low-molecular-weight form of occludin of 42 kDa upon NH3/
derived from urea metabolism by wild-type H. pylori resulted in a 20–30 % decrease in TER. After 24 h, the NH4Cl-treated cells showed a partial recovery of TER. In contrast, the control culture, or cultures that were exposed to supernatants derived from urease-deficient H. pylori, showed no significant decrease in TER. Occludin-specific immunoblots revealed the expression of a low-molecular-weight form of occludin of 42 kDa upon NH3/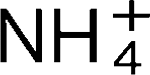 exposure. The results indicate that modulation of tight-junction function by H. pylori is ammonium-dependent and linked to the accumulation of a low-molecular-weight and detergent-soluble form of occludin.
exposure. The results indicate that modulation of tight-junction function by H. pylori is ammonium-dependent and linked to the accumulation of a low-molecular-weight and detergent-soluble form of occludin.
-
-
-
Functional analysis of EspB from enterohaemorrhagic Escherichia coli
More LessIn enterohaemorrhagic Escherichia coli (EHEC), the type III secretion protein EspB is translocated into the host cells and plays an important role in adherence, pore formation and effector translocation during infection. The secretion domain of EspB has been mapped previously. To define the other functional determinants of EspB, several plasmids encoding different fragments of EspB were created and analysed to see which of them lost the functions of the full-length molecule. One finding was that residues 118–190 of EspB were required for both efficient translocation of EspB and interaction of EspB with EspA. Additionally, the segment consisting of residues 217–312 was necessary for bacterial adherence. Furthermore, a predicted transmembrane domain (residues 99–118) was found to be critical for EHEC to cause red blood cell haemolysis, presumably by forming pores in the cell membrane. The same segment was also important for actin accumulation induced beneath the bacterial-attachment site. Taken together, these data indicate that the EspB protein (312 residues in total) has functions associated with its different regions. These regions may interact with each other or with other components of the type III system to orchestrate the intricate actions of EHEC during infection.
-
-
-
Functional analysis of the phospholipase C gene CaPLC1 and two unusual phospholipase C genes, CaPLC2 and CaPLC3, of Candida albicans
More LessPhospholipases C are known to be important regulators of cellular processes but may also act as virulence factors of pathogenic microbes. At least three genes in the genome of the human-pathogenic fungus Candida albicans encode phospholipases with conserved phospholipase C (Plc) motifs. None of the deduced protein sequences contain N-terminal signal peptides, suggesting that these phospholipases are not secreted. In contrast to its orthologue in Sacharomyces cerevisiae, CaPLC1 seems to be an essential gene. However, a conditional mutant with reduced transcript levels of CaPLC1 had phenotypes similar to Plc1p-deficient mutants in S. cerevisiae, including reduced growth on media causing increased osmotic stress, on media with a non-glucose carbon source, or at elevated or lower temperatures, suggesting that CaPlc1p, like the Plc1p counterpart in S. cerevisiae, may be involved in multiple cellular processes. Furthermore, phenotypic screening of the heterozygous ΔCaplc1/CaPLC1 mutant showed additional defects in hyphal formation. The loss of CaPLC1 cannot be compensated by two additional PLC genes of C. albicans (CaPLC2 and CaPLC3) encoding two almost identical phospholipases C with no counterpart in S. cerevisiae but containing structural elements found in bacterial phospholipases C. Although the promoter sequences of CaPLC2 and CaPLC3 differed dramatically, the transcriptional pattern of both genes was similar. In contrast to CaPLC1, CaPLC2 and CaPLC3 are not essential. Although Caplc2/3 mutants had reduced abilities to produce hyphae on solid media, these mutants were as virulent as the wild-type in a model of systemic infection. These data suggest that C. albicans contains two different classes of phospholipases C which are involved in cellular processes but which have no specific functions in pathogenicity.
-
-
-
Dimeric Brucella abortus Irr protein controls its own expression and binds haem
More LessBrucella abortus needs to synthesize haem in order to replicate intracellularly and to produce virulence in mice. Thus, to gain insight into the pathogenesis of the bacterium, regulatory proteins of the haem biosynthetic pathway were sought. An iron response regulator (Irr) from Bradyrhizobium japonicum, which is a close relative of Brucella, was previously described as being involved in the coordination of haem biosynthesis and iron availability. The Bru. abortus genome was searched for an irr orthologue gene, and the Bru. abortus irr gene was cloned, sequenced and disrupted. A null mutant was constructed that accumulated protoporphyrin IX under conditions of iron deprivation. This phenotype was overcome by a complementing plasmid carrying the wild-type irr. Purified recombinant Bru. abortus Irr behaved as a stable dimer and bound haem. Interestingly, in vivo, Irr was only detected in cells obtained from iron-limited cultures and the protein downregulated its own transcription. Through lacZ fusion, it was demonstrated that iron did not regulate irr transcription. The data reported show that Bru. abortus Irr is a homodimeric protein that is accumulated in iron-limited cells, controls its own transcription and downregulates the biosynthesis of haem precursors.
-
- Plant-Microbe Interactions
-
-
-
A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria
More LessA series of promoter probe vectors for use in Gram-negative bacteria has been made in two broad-host-range vectors, pOT (pBBR replicon) and pJP2 (incP replicon). Reporter fusions can be made to gfpUV, gfpmut3.1, unstable gfpmut3.1 variants (LAA, LVA, AAV and ASV), gfp+, dsRed2, dsRedT.3, dsRedT.4, mRFP1, gusA or lacZ. The two vector families, pOT and pJP2, are compatible with one another and share the same polylinker for facile interchange of promoter regions. Vectors based on pJP2 have the advantage of being ultra-stable in the environment due to the presence of the parABCDE genes. As a confirmation of their usefulness, the dicarboxylic acid transport system promoter (dctA p) was cloned into a pOT (pRU1097)- and a pJP2 (pRU1156)-based vector and shown to be expressed by Rhizobium leguminosarum in infection threads of vetch. This indicates the presence of dicarboxylates at the earliest stages of nodule formation.
-
-
- Theoretical Microbiology
-
-
-
Combining microarray and genomic data to predict DNA binding motifs
More LessThe ability to detect regulatory elements within genome sequences is important in understanding how gene expression is controlled in biological systems. In this work, microarray data analysis is combined with genome sequence analysis to predict DNA sequences in the photosynthetic bacterium Rhodobacter sphaeroides that bind the regulators PrrA, PpsR and FnrL. These predictions were made by using hierarchical clustering to detect genes that share similar expression patterns. The DNA sequences upstream of these genes were then searched for possible transcription factor recognition motifs that may be involved in their co-regulation. The approach used promises to be widely applicable for the prediction of cis-acting DNA binding elements. Using this method the authors were independently able to detect and extend the previously described consensus sequences that have been suggested to bind FnrL and PpsR. In addition, sequences that may be recognized by the global regulator PrrA were predicted. The results support the earlier suggestions that the DNA binding sequence of PrrA may have a variable-sized gap between its conserved block elements. Using the predicted DNA binding sequences, a whole-genome-scale analysis was performed to determine the relative importance of the interplay between the three regulators PpsR, FnrL and PrrA. Results of this analysis showed that, compared to the regulation by PpsR and FnrL, a much larger number of genes are candidates to be regulated by PrrA. The study demonstrates by example that integration of multiple data types can be a powerful approach for inferring transcriptional regulatory patterns in microbial systems, and it allowed the detection of photosynthesis-related regulatory patterns in R. sphaeroides.
-
-
Volumes and issues
-
Volume 170 (2024)
-
Volume 169 (2023)
-
Volume 168 (2022)
-
Volume 167 (2021)
-
Volume 166 (2020)
-
Volume 165 (2019)
-
Volume 164 (2018)
-
Volume 163 (2017)
-
Volume 162 (2016)
-
Volume 161 (2015)
-
Volume 160 (2014)
-
Volume 159 (2013)
-
Volume 158 (2012)
-
Volume 157 (2011)
-
Volume 156 (2010)
-
Volume 155 (2009)
-
Volume 154 (2008)
-
Volume 153 (2007)
-
Volume 152 (2006)
-
Volume 151 (2005)
-
Volume 150 (2004)
-
Volume 149 (2003)
-
Volume 148 (2002)
-
Volume 147 (2001)
-
Volume 146 (2000)
-
Volume 145 (1999)
-
Volume 144 (1998)
-
Volume 143 (1997)
-
Volume 142 (1996)
-
Volume 141 (1995)
-
Volume 140 (1994)
-
Volume 139 (1993)
-
Volume 138 (1992)
-
Volume 137 (1991)
-
Volume 136 (1990)
-
Volume 135 (1989)
-
Volume 134 (1988)
-
Volume 133 (1987)
-
Volume 132 (1986)
-
Volume 131 (1985)
-
Volume 130 (1984)
-
Volume 129 (1983)
-
Volume 128 (1982)
-
Volume 127 (1981)
-
Volume 126 (1981)
-
Volume 125 (1981)
-
Volume 124 (1981)
-
Volume 123 (1981)
-
Volume 122 (1981)
-
Volume 121 (1980)
-
Volume 120 (1980)
-
Volume 119 (1980)
-
Volume 118 (1980)
-
Volume 117 (1980)
-
Volume 116 (1980)
-
Volume 115 (1979)
-
Volume 114 (1979)
-
Volume 113 (1979)
-
Volume 112 (1979)
-
Volume 111 (1979)
-
Volume 110 (1979)
-
Volume 109 (1978)
-
Volume 108 (1978)
-
Volume 107 (1978)
-
Volume 106 (1978)
-
Volume 105 (1978)
-
Volume 104 (1978)
-
Volume 103 (1977)
-
Volume 102 (1977)
-
Volume 101 (1977)
-
Volume 100 (1977)
-
Volume 99 (1977)
-
Volume 98 (1977)
-
Volume 97 (1976)
-
Volume 96 (1976)
-
Volume 95 (1976)
-
Volume 94 (1976)
-
Volume 93 (1976)
-
Volume 92 (1976)
-
Volume 91 (1975)
-
Volume 90 (1975)
-
Volume 89 (1975)
-
Volume 88 (1975)
-
Volume 87 (1975)
-
Volume 86 (1975)
-
Volume 85 (1974)
-
Volume 84 (1974)
-
Volume 83 (1974)
-
Volume 82 (1974)
-
Volume 81 (1974)
-
Volume 80 (1974)
-
Volume 79 (1973)
-
Volume 78 (1973)
-
Volume 77 (1973)
-
Volume 76 (1973)
-
Volume 75 (1973)
-
Volume 74 (1973)
-
Volume 73 (1972)
-
Volume 72 (1972)
-
Volume 71 (1972)
-
Volume 70 (1972)
-
Volume 69 (1971)
-
Volume 68 (1971)
-
Volume 67 (1971)
-
Volume 66 (1971)
-
Volume 65 (1971)
-
Volume 64 (1970)
-
Volume 63 (1970)
-
Volume 62 (1970)
-
Volume 61 (1970)
-
Volume 60 (1970)
-
Volume 59 (1969)
-
Volume 58 (1969)
-
Volume 57 (1969)
-
Volume 56 (1969)
-
Volume 55 (1969)
-
Volume 54 (1968)
-
Volume 53 (1968)
-
Volume 52 (1968)
-
Volume 51 (1968)
-
Volume 50 (1968)
-
Volume 49 (1967)
-
Volume 48 (1967)
-
Volume 47 (1967)
-
Volume 46 (1967)
-
Volume 45 (1966)
-
Volume 44 (1966)
-
Volume 43 (1966)
-
Volume 42 (1966)
-
Volume 41 (1965)
-
Volume 40 (1965)
-
Volume 39 (1965)
-
Volume 38 (1965)
-
Volume 37 (1964)
-
Volume 36 (1964)
-
Volume 35 (1964)
-
Volume 34 (1964)
-
Volume 33 (1963)
-
Volume 32 (1963)
-
Volume 31 (1963)
-
Volume 30 (1963)
-
Volume 29 (1962)
-
Volume 28 (1962)
-
Volume 27 (1962)
-
Volume 26 (1961)
-
Volume 25 (1961)
-
Volume 24 (1961)
-
Volume 23 (1960)
-
Volume 22 (1960)
-
Volume 21 (1959)
-
Volume 20 (1959)
-
Volume 19 (1958)
-
Volume 18 (1958)
-
Volume 17 (1957)
-
Volume 16 (1957)
-
Volume 15 (1956)
-
Volume 14 (1956)
-
Volume 13 (1955)
-
Volume 12 (1955)
-
Volume 11 (1954)
-
Volume 10 (1954)
-
Volume 9 (1953)
-
Volume 8 (1953)
-
Volume 7 (1952)
-
Volume 6 (1952)
-
Volume 5 (1951)
-
Volume 4 (1950)
-
Volume 3 (1949)
-
Volume 2 (1948)
-
Volume 1 (1947)
Most Read This Month


