 -
Volume 70,
Issue 7,
1989
-
Volume 70,
Issue 7,
1989
Volume 70, Issue 7, 1989
- Review Article
-
-
-
Biology and Pathogenesis of Lentiviruses
More LessLentiviruses are a family of retroviruses linked by similarities in genetic composition, molecular mechanisms of replication and in biological interactions with their hosts. They are best known as agents of slow disease syndromes that begin insidiously after prolonged periods of subclinical infection and progress slowly leading to the degeneration of multiple organ systems, cachexia and death. The viruses are species-specific in host range and several have been recognized as pathogens of domestic animals, non-human primates and humans. The prototypes of the family are the agents causing maedi-visna in sheep and infectious anaemia in horses. These diseases have been known for several decades and studies on the biology of the viruses have provided a fund of information that predicted most of the properties of their human counterpart which was identified only 6 years ago as the aetiological agent of AIDS.
Lentiviruses persist indefinitely in their hosts and replicate continuously at variable rates during the course of the lifelong infection. Persistent replication of the viruses depends on their ability to circumvent host defences. In this respect the agents have evolved a repertoire of strategies that surpass those of any other known pathogen. Studies on immunization have shown that the viruses are poor immunogens for the induction of protective antibodies but this varies among the virus families. The ‘Achilles heel’ of these viruses lies in their absolute dependence on blood and tissue fluids for host-to-host transmission.
The viruses are tropic for macrophages in vivo and replication is regulated by non-structural viral genes and by factors produced by the activated host cells. Clinical disease is related to virus replication in macrophages and two types of disease are produced: primary disease, caused directly by the lentivirus and secondary disease, caused by opportunistic agents.
Primary disease is the major pathological manifestation of infection in domestic animals and is associated with the activation of virus replication in selective populations of macrophages which are tissue-specific. Host and viral factors determine which populations will be involved in the support of virus replication. Primary disease also occurs in humans and is exemplified by the unique lesions in the brain, lungs and lymph nodes of patients with AIDS and AIDS-related complex. Development of primary lesions is associated with the enhanced production of cytokines that are produced partly by the macrophage and partly by the lymphocytes, responding to antigens presented by the immunologically activated, infected macrophages.
Secondary disease (AIDS) is caused by opportunistic agents which proliferate as a result of the loss of function of activated helper T lymphocytes. In humans, macaques and cats, these cells are highly susceptible to lysis by the respective viruses and/or by virus-infected macrophages under cell culture conditions. Loss of the lymphocytes in vivo leads to profound immunosuppression. Helper T lymphocytes of ungulate animals are not as sensitive to the specific lentivirus infecting the animal and this correlates with the lack of secondary diseases in these animals.
-
-
- Animal
-
-
-
Asymptomatic Infection of the Central Nervous System by the Macaque Immunosuppressive Type D Retrovirus, SRV-1
More LessSUMMARYThe aetiological agent of spontaneously occurring simian acquired immune deficiency syndrome (SAIDS) in rhesus monkeys (Macaca mulatta) at the California Primate Research Center is a type D retrovirus designated SAIDS retrovirus serotype 1 (SRV-1). SRV-1 DNA and RNA have previously been detected in the brains of rhesus monkeys with SAIDS in the absence of viral antigen or neuropathological lesions. In this study we further define the relationship between SRV-1 and the central nervous system (CNS) in rhesus monkeys by examining the CNS for infectious SRV-1, viral antigen and anti-SRV-1 antibodies. In addition, cerebrospinal fluid (CSF) was assayed for alterations in IgG and albumin levels, IgG/albumin ratios and cell count in comparison to uninfected control animals. No differences in CSF parameters were detected between infected and uninfected animals except for the presence of infectious SRV-1 which was isolated from the CSF from 13 out of 19 (68%) viraemic rhesus monkeys. The probable source of this virus was the choroid plexus, where approximately 1 in 1000 surface epithelial cells were found to contain viral antigen by immunohistochemistry. Antibodies against SRV-1 were not detected in the CSF even when present in the serum. Neither infectious virus nor viral antigen were found in the brain parenchyma of any animal examined. Thus infection of the CNS by SRV-1 appears to be subclinical without an intrathecal immune response. This may be related to the apparent restriction of productive infection in the CNS to cells of the choroid plexus.
-
-
-
-
A Human Melanoma Cell Line Highly Susceptible to Influenza C Virus
More LessSUMMARYThe relative amounts of influenza C virus-specific receptors of 25 established lines of mammalian cells including four lines of human malignant melanoma origin were compared by virus binding experiments. All the human melanoma cell cultures studied possessed two to four times more receptors than were found on MDCK cells, a cell line known to be highly susceptible to influenza C virus. It may therefore be a feature common to human melanoma cells that O-acetylsialic acid, a determinant for the attachment of influenza C virus, exists in large quantities on their surface. This is not specific to melanoma cells, however, since several human cell lines derived from lung cancer, gastric cancer, and placenta specimens also exhibited high levels of virus binding. Twenty of 25 virus-binding cell cultures were further examined for their ability to support the replication of influenza C virus. In the presence of trypsin (5 to 20 μg/ml), the virus was found to undergo multiple cycles of replication much more efficiently in the HMV-II line of human melanoma cells than in MDCK cells. Additionally, by using HMV-II cells as a host, we succeeded in isolating two influenza C strains (C/Yamagata/1/88, C/Yamagata/2/88) from 241 throat swabs collected from patients with acute respiratory illness.
-
-
-
Bluetongue Virus Infection of Bovine Monocytes
More LessSUMMARYCultures of adherent and non-adherent bovine blood mononuclear cells were infected with bluetongue virus (BTV) serotype 10. Production of BTV proteins in mononuclear cell cultures was detected by immune precipitation of viral proteins from [35S]methionine-labelled extracts of these cells, by immunofluorescence staining of cells using monoclonal antibodies (MAbs) to BTV proteins VP7 and NS2, and by flow cytometry with MAbs to VP2, VP7, NS1 and NS2. BTV-infected cells were most numerous in cultures of adherent mononuclear cells; infected cells were initially identified as monocytes on the basis of their morphology, and size and scatter characteristics as determined by analysis with a fluorescence-activated cell sorter (FACS). The majority of adherent mononuclear cells with these scatter characteristics were confirmed to be monocytes by FACS analysis with a MAb specific for bovine monocytes. Identification of BTV-infected adherent mononuclear cells as monocytes was further established by double immunofluorescent labelling, as infected adherent cells reacted with the MAb specific for bovine monocytes, and with another MAb specific for class II antigen. Infection of adherent mononuclear cells was also confirmed by transmission electron microscopy, as BTV virions and tubules were present in lysates of cultures of BTV-infected adherent mononuclear cells and within the cytoplasm of adherent cells. In contrast, BTV proteins were detected in few cells identified as lymphocytes on the basis of their scatter characteristics, and mean fluorescence of such cells was considerably less than that of BTV-infected monocytes. Viraemia persisted until 35 days after inoculation of a colostrum-deprived calf inoculated with BTV. Virus was isolated from blood mononuclear cells at 1 week after infection of the calf, but not thereafter. BTV infection of blood mononuclear cells was demonstrated until 9 days after inoculation by indirect immunofluorescence staining of mononuclear cells. In contrast, virus was consistently isolated from erythrocyte-enriched preparations throughout viraemia in titres comparable to those in whole blood. These results indicate that although bovine monocytes are readily infected in vitro with this strain of BTV serotype 10, infection of blood monocytes is unlikely to be responsible for the prolonged viraemia that consistently occurs in BTV-infected cattle.
-
-
-
Completion of the Sequence of Bluetongue Virus Serotype 10 by the Characterization of a Structural Protein, VP6, and a Non-structural Protein, NS2
More LessA. Fukusho, Y. Yu, S. Yamaguchi and P. RoySUMMARYThe sequence of cDNA clones representing the entire genome of bluetongue virus serotype 10 (BTV-10) has been completed by the analysis of data obtained for the S8 and S9 segments. Each DNA clone has been sequenced completely and the deduced amino acid sequences have been analysed. The sequences of the S8 and S9 gene products as well as another two previously published small gene products (S7 and S10) have been compared with the corresponding size gene products of reovirus type 1. The data do not indicate a relationship between the small proteins of these two viruses except some distant homologies between the BTV VP7 protein and the σ1 protein of reovirus. The characteristics of all the BTV-10 genome segments, the deduced primary gene products and their possible functions are summarized.
-
-
-
Genotypic Selection Following Coinfection of Cultured Cells with Subgroup 1 and Subgroup 2 Human Rotaviruses
More LessSUMMARYThe purpose of this study was to determine why identifiable reassortants between subgroup 1 and subgroup 2 rotaviruses have been so rarely isolated from human specimens. Cultured cells were coinfected with pairs of subgroup 1 and 2 human rotaviruses and passaged multiple times to simulate natural reassortant formation and selection in vivo. After coinfection of MA-104 cells with subgroup 1 (DS-1) and subgroup 2 (either Wa or P) strains, approximately 14% of the plaque-picked progeny were shown to be reassortants. During multiple passages of these coinfected cultures, however, complete (Wa virus coinfection) or nearly complete (P virus coinfection) loss of detectable DS-1 segments from progeny was observed. Thus, when all segments of the subgroup 2 viruses were present in coinfected cultures, these segments dominated in the selected progeny. Coinfection with subgroup 1-subgroup 2 rotavirus reassortants and the DS-1 strain followed by multiple passages, however, resulted in complete loss of some segments from the subgroup 2 strains originally present in the reassortants. Therefore, segments from the parental subgroup 2 viruses appeared to be selected in toto during multiple passages because they were dominant as a group, not because individual segments of these viruses were consistently favoured over their subgroup 1 virus counterparts.
-
-
-
Genetic Relatedness among Structural Protein Genes of Dengue 1 Virus Strains
More LessSUMMARYThe structural protein-coding genomic regions of dengue virus type 1 (DEN-1) strains representing three distinct topotypes (Thailand, Philippines and Caribbean) were cloned and sequenced. In addition the envelope (E) nucleotide sequences of two recent Caribbean topotype DEN-1 isolates were obtained by direct RNA sequencing. The nucleotide sequence of the DEN-1 viruses in the structural gene region was found to be highly conserved with greater than 95% nucleotide sequence homology and with less than 4% change in the amino acid sequence. Although there was a less than 2% change in the nucleotide sequence of DEN-1 E proteins, strains could be differentiated by the clusters of nucleotide changes. Furthermore, the deduced amino acid changes in the E protein were clustered primarily within the proposed immunologically reactive regions. Genomic nucleotide sequence comparisons did not define geographical or virulence markers but located unique clusters of nucleotide/amino acid changes for each of the three topotypes of DEN-1 viruses examined.
-
-
-
Several Rat Cell Lines Share a Common Defect in Their Inability to Internalize Murine Coronaviruses Efficiently
More LessSUMMARYInfection of rat cells, Schwannoma RN2, hepatoma HTC or myoblast L6, with the murine coronavirus JHM strain results in a persistent infection characterized by the release of virus over an extended period of time with a limited cytopathology. Several stages of the viral replication cycle have been examined in these cells in comparison to those in mouse L2 cells, which are totally permissive to JHM infection. Although the rat cells bound as much virus as the mouse cells their ability to internalize it was 40-fold less efficient than the mouse cells. This lower internalization efficiency was not enhanced by pH shock of infected cells, but was by treatment with polyethylene glycol. In all cell types there appeared to be no major differences in the ability of the internalized virus to replicate the viral RNA as determined by slot-blot analysis with a radiolabelled viral cDNA. A similar genetic mechanism appears to be operative in the lines because somatic cell hybrids formed between these lines in various combinations were also deficient in the ability to internalize bound virus. Taken together these results imply that rat cell lines in general share a common deficiency in their inability to internalize murine coronaviruses efficiently. This low efficiency in viral internalization may explain in part the ability of these lines to sustain persistent infections.
-
-
-
Antigenic and Polypeptide Structure of Turkey Enteric Coronaviruses as Defined by Monoclonal Antibodies
More LessSUMMARYTwenty-nine hybridoma cell lines, producing monoclonal antibodies (MAbs) to the Minnesota strain of turkey enteric coronavirus (TCV), have been established by fusion of Sp2/0 myeloma cells with spleen cells from BALB/c mice immunized with purified preparations of the egg-adapted or tissue culture-adapted virus. The hybridomas produced mainly IgG2a or IgG1 antibodies. Western immunoblotting experiments with purified virus, and immunoprecipitation tests with [35S]methionine-labelled infected cell extracts, allowed assessment of the polypeptide specificity of the MAbs. Sixteen hybridomas secreted antibodies directed to the peplomeric protein (E2, gp200/gp100) and putative intracellular precursors of apparent M r 170K to 180K and 90K. Four hybridomas produced antibodies that selectively reacted with a glycoprotein with an M r of 140K (E3). This polypeptide species corresponded to the major structural component of small granular projections, located near the base of the larger bulbous peplomers, and was found to be responsible for haemagglutination. The major neutralization-mediating determinants were found to be carried by both E2 and E3 glycoproteins. Eight hybridomas produced MAbs directed to the major nucleocapsid protein (N, 52K), and only one MAb reacted with a low M r structural glycoprotein (24K), corresponding to the matrix (El) protein. By indirect immunofluorescence, MAbs of different specificity also revealed distinct patterns of distribution of the viral antigens within the cells. The location on the virion of the antigenic determinants recognized by MAbs of different specificity was determined by the use of an immunogold electron microscopy technique. Comparison of nine TCV Quebec strains, using MAbs directed to peplomer and haemagglutinin proteins of the prototype Minnesota strain, confirmed their close antigenic relationship, but also revealed the occurrence of at least two distinct antigenic groups.
-
-
-
Proteins Specified by Bovine Herpesvirus Type 4: Structural Proteins of the Virion and Identification of Two Major Glycoproteins by Using Monoclonal Antibodies
More LessSUMMARYBovine herpesvirus type 4 proteins were identified by PAGE of [35S]methionine- or [3H]glucosamine-labelled purified virions. Thirty-one monoclonal antibodies (MAbs) raised against the V. Test strain were used to identify 29 proteins, ten of which were glycosylated. All of these glycoproteins belonged to the viral envelope and a 140K nonglycosylated protein appeared to be the major nucleocapsid protein. The MAbs were classified into two groups. The first group precipitated three glycoproteins of M r 150K, 120K and 51K. The 120K and 51K glycoproteins were linked by disulphide bonds and the 150K glycoprotein was linked to the others by non-covalent bonds. The second group precipitated a different 120K glycoprotein.
-
-
-
Epstein-Barr Virus Latent Gene Expression during the Initiation of B Cell Immortalization
More LessSUMMARYEpstein-Barr virus (EBV) has the capacity to immortalize a subpopulation of resting B lymphocytes. Lymphoblastoid cell lines (LCL) established in this way carry the latent EBV genome as multiple copies of an extrachromasomal episome. Viral gene expression in LCLs is highly restricted; products identified correspond to a membrane protein (latent membrane protein; LMP), a nuclear antigen complex (Epstein-Barr nuclear antigens; EBNAs 1 to 6), two small RNA species (EBERs 1 and 2) and RNA species thought to encode a second membrane-associated polypeptide designated terminal protein (TP). Here we have investigated the temporal sequence of expression of the characterized ‘latent’ proteins during the initiation of immortalization when resting B cells are stimulated to enter and traverse the cell cycle. The analysis has been carried out on prolymphocytic leukaemia cells infected in vitro with either the immortalizing B95-8 strain of virus or the non-immortalizing P3HR1 strain. The results reveal that following B95-8 infection, a sequence of EBV expression is initiated within approximately 8 h with the synthesis of detectable levels of EBNA 2 shortly followed by EBNAs 1, 3, 4, 5 and 6. There is then a delay of approximately 40 h until the expression of LMP completes the latent pattern of proteins found in LCLs. P3HR1 infection, however, produces only transient expression of some EBNA species in a small percentage of cells after approximately 48 h. These observations suggest the failure of P3HR1 virus to immortalize may not be due solely to the absence of EBNA 2 expression and that cellular and/or virus-mediated events occur after EBNA synthesis which then facilitate efficient LMP expression and immortalization.
-
-
-
Effects of L3T4+ Lymphocyte Depletion on Acute Murine Cytomegalovirus Infection
More LessSUMMARYWe examined the role ofT lymphocytes bearing the L3T4 phenotype in acute murine cytomegalovirus (MCMV) infection. In vivo administration of rat IgG2b monoclonal antibody (MAb) GK 1.5 was used to deplete mice of L3T4+ lymphocytes during acute MCMV infection. Unlike the saline-treated controls that resolved their infections, mice receiving the MAb developed persistent and high levels of virus in the salivary gland, lung and spleen. The production of antibody to MCMV was delayed and the titres achieved were markedly less than in the controls. Despite the higher levels of virus replication, there was no increase in mortality seen in animals treated with the MAb. Following intraperitoneal challenge with MCMV, depletion of L3T4+ lymphocytes was protective, increasing the dose of MCMV required to produce death. These data indicate that T lymphocytes of the L3T4 phenotype influence the degree of MCMV replication during acute infection and may contribute to mortality following intraperitoneal virus challenge.
-
-
-
Effects of Acyclovir on Herpes Simplex Virus Type 1 Infection in Mice Treated with 12-O-Tetradecanoylphorbol 13-acetate
More LessSUMMARYThe purpose of this study was to determine whether infectious herpes simplex virus type L (HSV-1) has tumorigenic properties and, if so, whether inhibition of the cytolytic replicative cycle of the virus after infection enhances tumour development. Eighty mice were subjected to repeated inoculation of HSV-1 on their upper lips after scarification, and systemic administration of acyclovir (ACV). 12-O-tetradecanoylphorbol 13-acetate (TPA) was used as the tumour promoter. The tumour incidence was compared to control groups each of 40 mice that were either not treated with ACV, not treated with TPA, not infected with HSV or only scarified. In the virus-infected group treated with ACV and TPA, 25% of the animals developed tumours. In the HSV-infected group treated with TPA only, 25% of the animals also developed tumours. The uninfected animals which were not treated with TPA developed tumours to a significantly lesser degree. In conclusion, the combined effects of HSV-1 and TPA, with or without ACV treatment, resulted in a significant increase in the number of tumours in comparison to the control groups.
-
-
-
Quantification of Herpes Simplex Virus Infection in Cervical Ganglia of Mice
More LessSUMMARYMice were inoculated with herpes simplex virus (HSV) type 1 in the skin of the neck. The extent of primary and latent infection in the second and third cervical ganglia was investigated. Immunoperoxidase staining of ganglia during primary infection demonstrated HSV antigens initially in a restricted area of the ganglion. By the 5th day after infection, antigen was more widespread. Such a change in the staining pattern is explicable in terms of the zosteriform spread of virus from neurons innervating the site of infection to others supplying other areas of the dermatome. A maximum of approximately 10% of neurons became infected. By the 7th day staining was limited to a few cells. During latent infection, enzymic disaggregation of ganglia followed by immunoperoxidase staining or infectious centre assay indicated that virus reactivation began within 30 h of removal of ganglia and occurred in approximately 1% of viable neurons.
-
-
-
Nucleotide Sequence and Characterization of the Marek’s Disease Virus Homologue of Glycoprotein B of Herpes Simplex Virus
More LessSUMMARYThe Marek’s disease virus (MDV) homologue of the herpes simplex virus (HSV) gene encoding glycoprotein B (gB) has been identified within BamHI fragments I3 and K3 of the ‘highly oncogenic’ strain RB1B of MDV. The entire nucleotide sequence of the gene has been determined and its predicted amino acid sequence shown to share gross overall structural features with the gB genes of HSV, varicella-zoster virus (VZV) and other mammalian herpesviruses. In particular, all 10 cysteine residues were conserved in MDV gB and there was extensive homology throughout the gene with VZV, HSV and pseudorabies virus except for the N and C termini. The overall percentage amino acid identity between MDV gB and gB of the alphaherpesviruses had a mean of 50% which was almost twice that between cytomegalovirus and Epstein-Barr virus. Northern blot analysis showed that the main RNA transcribed from this gene is approx. 2·7 kb in size. Antibodies raised against synthetic peptides (residues 250 to 271 and 304 to 330) allowed the identification of a family of serologically related glycoproteins of M r 110K, 64K and 48K in extracts of MDV-infected cells using immunoblots. Furthermore, the antisera were able to differentiate between the antigens of MDV and herpesvirus of turkeys in immunoblots. Immunofluorescence tests indicated that MDV gB is associated with granules in the cytoplasm and is present at the surface of MDV-infected cells.
-
-
-
Expression of Glycoprotein D of Herpes Simplex Virus Type 1 in a Recombinant Baculovirus: Protective Responses and T Cell Recognition of the Recombinant-infected Cell Extracts
More LessSUMMARYRecombinant baculoviruses expressing glycoprotein D (gD) of herpes simplex virus type 1 (HSV-1) have been generated. The proteins expressed from the recombinants have been characterized using monoclonal antibodies on Western blots and by immunoprecipitation. Partially glycosylated 48K polypeptides have been identified as products of the gD gene. Polyclonal sera from H-2k mice infected with HSV-1 recognized the same polypeptides. Furthermore, draining lymph node cells from H-2k mice infected with HSV-1 proliferated in vitro in response to recombinant-infected cell extracts. Immunization with such extracts generated high titre complement-dependent and -independent neutralizing antibody and the mice were protected against a challenge with HSV-1.
-
-
-
Characterization of the Genetic Organization of the HindIII M Region of the Multicapsid Nuclear Polyhedrosis Virus of Orgyia pseudotsugata Reveals Major Differences among Baculoviruses
More LessSUMMARYA region including the 4 kb HindIII M fragment of the multicapsid nuclear polyhedrosis virus (MNPV) of Orgyia pseudotsugata (OpMNPV) genome was sequenced, transcriptionally mapped, and compared to the homologous region in the MNPV of Autographa californica (AcMNPV). Five open reading frames (ORFs) oriented in the same direction were identified and were found to be expressed at late times post-infection. The mRNAs from the five ORFs were found to coterminate at a single site downstream of ORF 5. The conserved late gene promoter/mRNA start site sequence (
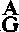 TAAG) was present upstream of all the ORFs, but did not appear to be the major site of mRNA initiation for two of the ORFs as determined by primer extension analysis. These data indicated that use of this signal for transcriptional initiation may vary between different ORFs. The predicted amino acid sequences for the five ORFs of AcMNPV and OpMNPV were compared and amino acid homologies of 26 to 72% were observed. The comparison revealed a number of major differences in the genomes of the two viruses. A putative transposable element of 634 nucleotides was found to be inserted into the previously reported AcMNPV ORF 1 sequence. In addition, it was found that a region corresponding to the 4 kb HindIII K/EcoRI S/hr5 region of AcMNPV was not present in the OpMNPV genome.
TAAG) was present upstream of all the ORFs, but did not appear to be the major site of mRNA initiation for two of the ORFs as determined by primer extension analysis. These data indicated that use of this signal for transcriptional initiation may vary between different ORFs. The predicted amino acid sequences for the five ORFs of AcMNPV and OpMNPV were compared and amino acid homologies of 26 to 72% were observed. The comparison revealed a number of major differences in the genomes of the two viruses. A putative transposable element of 634 nucleotides was found to be inserted into the previously reported AcMNPV ORF 1 sequence. In addition, it was found that a region corresponding to the 4 kb HindIII K/EcoRI S/hr5 region of AcMNPV was not present in the OpMNPV genome.
-
-
-
Physical Map of Anticarsia gemmatalis Nuclear Polyhedrosis Virus (AgMNPV-2) DNA
More LessSUMMARYA physical map for a plaque-purified isolate of a baculovirus which infects Anticarsia gemmatalis Hübner (velvetbean caterpillar), was constructed. By applying multiple-enzyme digestion and DNA–DNA hybridization techniques to the viral DNA restriction fragments, a total of 51 restriction sites were mapped. This baculovirus was found to possess a 133 kb circular dsDNA genome typical of members of baculovirus subgroup A.
-
-
-
Nucleotide Sequence of the Polyhedrin Gene of Bombyx mori Cytoplasmic Polyhedrosis Virus A Strain with Nuclear Localization of Polyhedra
More LessSUMMARYBombyx mori cytoplasmic polyhedrosis virus (BmCPV) strain H produces many hexahedral polyhedra (inclusion bodies) in the cytoplasm of insect midgut epithelial cells. The mutant A strain, however, produces polyhedra in the nucleus. We determined cDNA sequences of the polyhedrin genes, the smallest of the 10 genome segments, of these two strains. The polyhedrin genes of the H and A strains were 944 bp long, and encoded polypeptides of 248 amino acids (M r 28500) and 252 amino acids (M r 29000), respectively. The extra four amino acid residues at the carboxy terminus of the strain A polyhedrin (Arg-Leu-Leu-Val) were the result of a single nucleotide substitution at an opal stop codon (TGA → CGA). A further amino acid substitution of the histidine residue at position 101 (His → Tyr) was seen. The carboxy-terminal extension revealed a considerable similarity to the consensus amino acid sequence of the DNA-binding domain of many DNA-binding proteins. We discuss the relationship between the intracellular localization of polyhedrins and mutations in their amino acid sequences.
-
-
-
Use of a Monoclonal Antibody Specific for Wild-type Yellow Fever Virus to Identify a Wild-type Antigenic Variant in 17D Vaccine Pools
More LessSUMMARYMonoclonal antibodies (MAbs) against the Asibi wild-type strain of yellow fever (YF) virus were prepared and characterized. One of the MAbs (designated MAb 117) was shown, by cross-immunofluorescence tests with flaviviruses, to be specific for wild-type YF virus. This MAb was used in indirect immunofluorescence tests to identify wild-type antigenic variants in several different YF vaccine pools. Simultaneously, a vaccine-specific MAb prepared previously (MAb 864) was used to identify YF strain 17D vaccine type variants in the wild-type Asibi virus preparation. One variant, isolated by plaque purification from a 17D vaccine pool, possessed the wild-type epitope and was neurovirulent in infant mice whereas other variants, lacking the wild-type epitope but with vaccine-specific epitopes (identified by MAb 411), were avirulent in infant mice. Avirulent variants were able to infect mice and induce antibody. Virus-specific antigen was still detected in the brains of these mice 4 weeks after inoculation, suggesting that persistent infections were developing. These results demonstrate the antigenic heterogeneity of 17D vaccine preparations. They also point to the potential risk of selection of wild-type variants in YF vaccine preparations and re-emphasize the need for modernization of techniques and more effective control measures to be taken during the production of YF vaccine.
-
Volumes and issues
-
Volume 105 (2024)
-
Volume 104 (2023)
-
Volume 103 (2022)
-
Volume 102 (2021)
-
Volume 101 (2020)
-
Volume 100 (2019)
-
Volume 99 (2018)
-
Volume 98 (2017)
-
Volume 97 (2016)
-
Volume 96 (2015)
-
Volume 95 (2014)
-
Volume 94 (2013)
-
Volume 93 (2012)
-
Volume 92 (2011)
-
Volume 91 (2010)
-
Volume 90 (2009)
-
Volume 89 (2008)
-
Volume 88 (2007)
-
Volume 87 (2006)
-
Volume 86 (2005)
-
Volume 85 (2004)
-
Volume 84 (2003)
-
Volume 83 (2002)
-
Volume 82 (2001)
-
Volume 81 (2000)
-
Volume 80 (1999)
-
Volume 79 (1998)
-
Volume 78 (1997)
-
Volume 77 (1996)
-
Volume 76 (1995)
-
Volume 75 (1994)
-
Volume 74 (1993)
-
Volume 73 (1992)
-
Volume 72 (1991)
-
Volume 71 (1990)
-
Volume 70 (1989)
-
Volume 69 (1988)
-
Volume 68 (1987)
-
Volume 67 (1986)
-
Volume 66 (1985)
-
Volume 65 (1984)
-
Volume 64 (1983)
-
Volume 63 (1982)
-
Volume 62 (1982)
-
Volume 61 (1982)
-
Volume 60 (1982)
-
Volume 59 (1982)
-
Volume 58 (1982)
-
Volume 57 (1981)
-
Volume 56 (1981)
-
Volume 55 (1981)
-
Volume 54 (1981)
-
Volume 53 (1981)
-
Volume 52 (1981)
-
Volume 51 (1980)
-
Volume 50 (1980)
-
Volume 49 (1980)
-
Volume 48 (1980)
-
Volume 47 (1980)
-
Volume 46 (1980)
-
Volume 45 (1979)
-
Volume 44 (1979)
-
Volume 43 (1979)
-
Volume 42 (1979)
-
Volume 41 (1978)
-
Volume 40 (1978)
-
Volume 39 (1978)
-
Volume 38 (1978)
-
Volume 37 (1977)
-
Volume 36 (1977)
-
Volume 35 (1977)
-
Volume 34 (1977)
-
Volume 33 (1976)
-
Volume 32 (1976)
-
Volume 31 (1976)
-
Volume 30 (1976)
-
Volume 29 (1975)
-
Volume 28 (1975)
-
Volume 27 (1975)
-
Volume 26 (1975)
-
Volume 25 (1974)
-
Volume 24 (1974)
-
Volume 23 (1974)
-
Volume 22 (1974)
-
Volume 21 (1973)
-
Volume 20 (1973)
-
Volume 19 (1973)
-
Volume 18 (1973)
-
Volume 17 (1972)
-
Volume 16 (1972)
-
Volume 15 (1972)
-
Volume 14 (1972)
-
Volume 13 (1971)
-
Volume 12 (1971)
-
Volume 11 (1971)
-
Volume 10 (1971)
-
Volume 9 (1970)
-
Volume 8 (1970)
-
Volume 7 (1970)
-
Volume 6 (1970)
-
Volume 5 (1969)
-
Volume 4 (1969)
-
Volume 3 (1968)
-
Volume 2 (1968)
-
Volume 1 (1967)
Most Read This Month


